2JGZ
 
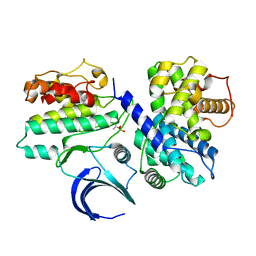 | | Crystal structure of phospho-CDK2 in complex with Cyclin B | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, G2/MITOTIC-SPECIFIC CYCLIN-B1 | | Authors: | Brown, N.R, Petri, E, Lowe, E.D, Skamnaki, V, Johnson, L.N. | | Deposit date: | 2007-02-17 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cyclin B and cyclin A confer different substrate recognition properties on CDK2.
Cell Cycle, 6, 2007
|
|
2JK1
 
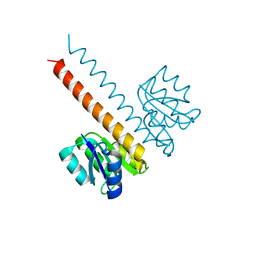 | | Crystal structure of the wild-type HupR receiver domain | | Descriptor: | HYDROGENASE TRANSCRIPTIONAL REGULATORY PROTEIN HUPR1, MAGNESIUM ION | | Authors: | Davies, K.M, Lowe, E.D, Venien-Bryan, C, Johnson, L.N. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Hupr Receiver Domain Crystal Structure in its Nonphospho and Inhibitory Phospho States.
J.Mol.Biol., 385, 2009
|
|
1H1Q
 
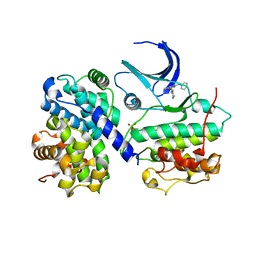 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU6094 | | Descriptor: | 2-ANILINO-6-CYCLOHEXYLMETHOXYPURINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of a Potent Purine-Based Cyclin-Dependent Kinase Inhibitor
Nat.Struct.Biol., 9, 2002
|
|
1H1S
 
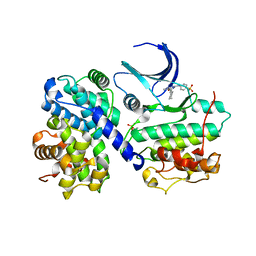 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU6102 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, O6-CYCLOHEXYLMETHOXY-2-(4'-SULPHAMOYLANILINO) PURINE | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of a Potent Purine-Based Cyclin-Dependent Kinase Inhibitor
Nat.Struct.Biol., 9, 2002
|
|
1H0V
 
 | | Human cyclin dependent protein kinase 2 in complex with the inhibitor 2-Amino-6-[(R)-pyrrolidino-5'-yl]methoxypurine | | Descriptor: | 5-{[(2-AMINO-9H-PURIN-6-YL)OXY]METHYL}-2-PYRROLIDINONE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Gibson, A.E, Arris, C.E, Bentley, J, Boyle, F.T, Curtin, N.J, Davies, T.G, Endicott, J.A, Golding, B.T, Grant, S, Griffin, R.J, Jewsbury, P, Johnson, L.N, Mesguiche, V, Newell, D.R, Noble, M.E.M, Tucker, J.A, Whitfield, H.J. | | Deposit date: | 2002-06-27 | | Release date: | 2003-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Ribose-Binding Domain of Cyclin-Dependent Kinases 1 and 2 with O(6)-Substituted Guanine Derivatives
J.Med.Chem., 45, 2002
|
|
1H1P
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU2058 | | Descriptor: | 6-O-CYCLOHEXYLMETHYL GUANINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of a Potent Purine-Based Cyclin-Dependent Kinase Inhibitor
Nat.Struct.Biol., 9, 2002
|
|
1H1R
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU6086 | | Descriptor: | 6-CYCLOHEXYLMETHOXY-2-(3'-CHLOROANILINO) PURINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of a potent purine-based cyclin-dependent kinase inhibitor.
Nat. Struct. Biol., 9, 2002
|
|
1H0W
 
 | | Human cyclin dependent protein kinase 2 in complex with the inhibitor 2-Amino-6-[cyclohex-3-enyl]methoxypurine | | Descriptor: | 1-AMINO-6-CYCLOHEX-3-ENYLMETHYLOXYPURINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Gibson, A.E, Arris, C.E, Bentley, J, Boyle, F.T, Curtin, N.J, Davies, T.G, Endicott, J.A, Golding, B.T, Grant, S, Griffin, R.J, Jewsbury, P, Johnson, L.N, Mesguiche, V, Newell, D.R, Noble, M.E.M, Tucker, J.A, Whitfield, H.J. | | Deposit date: | 2002-06-27 | | Release date: | 2003-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the ATP Ribose-Binding Domain of Cyclin-Dependent Kinases 1 and 2 with O(6)-Substituted Guanine Derivatives
J.Med.Chem., 45, 2002
|
|
1L5V
 
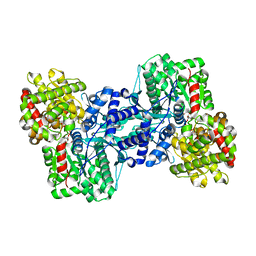 | | Crystal Structure of the Maltodextrin Phosphorylase complexed with Glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MALTODEXTRIN PHOSPHORYLASE, ... | | Authors: | Geremia, S, Campagnolo, M, Schinzel, R, Johnson, L.N. | | Deposit date: | 2002-03-08 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase
J.Mol.Biol., 322, 2002
|
|
1L5W
 
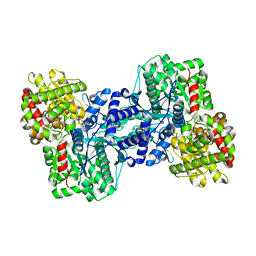 | | Crystal Structure of the Maltodextrin Phosphorylase Complexed with the Products of the Enzymatic Reaction between Glucose-1-phosphate and Maltotetraose | | Descriptor: | MALTODEXTRIN PHOSPHORYLASE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Geremia, S, Campagnolo, M, Schinzel, R, Johnson, L.N. | | Deposit date: | 2002-03-08 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase
J.Mol.Biol., 322, 2002
|
|
1L6I
 
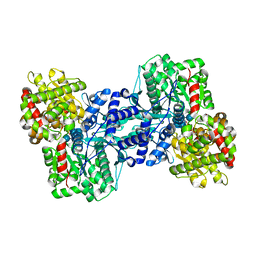 | | Crystal Structure of the Maltodextrin Phosphorylase complexed with the products of the enzymatic reaction between glucose-1-phosphate and maltopentaose | | Descriptor: | MALTODEXTRIN PHOSPHORYLASE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Geremia, S, Campagnolo, M, Schinzel, R, Johnson, L.N. | | Deposit date: | 2002-03-11 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase
J.Mol.Biol., 322, 2002
|
|
1TAH
 
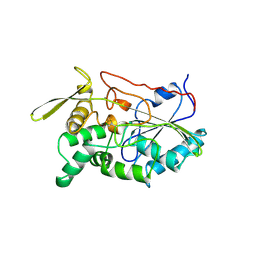 | | THE CRYSTAL STRUCTURE OF TRIACYLGLYCEROL LIPASE FROM PSEUDOMONAS GLUMAE REVEALS A PARTIALLY REDUNDANT CATALYTIC ASPARTATE | | Descriptor: | CALCIUM ION, LIPASE | | Authors: | Noble, M.E.M, Cleasby, A, Johnson, L.N, Egmond, M, Frenken, L.G.J. | | Deposit date: | 1993-12-21 | | Release date: | 1994-05-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate.
FEBS Lett., 331, 1993
|
|
1UA2
 
 | | Crystal Structure of Human CDK7 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein kinase 7 | | Authors: | Lolli, G, Lowe, E.D, Brown, N.R, Johnson, L.N. | | Deposit date: | 2004-08-11 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The Crystal Structure of Human CDK7 and Its Protein Recognition Properties
Structure, 12, 2004
|
|
1W98
 
 | | The structural basis of CDK2 activation by cyclin E | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, G1/S-SPECIFIC CYCLIN E1 | | Authors: | Lowe, E.D, Honda, R, Dubinina, E, Skamnaki, V, Cook, A, Johnson, L.N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of Cyclin E1/Cdk2: Implications for Cdk2 Activation and Cdk2-Independent Roles
Embo J., 24, 2005
|
|
2BWF
 
 | | Crystal structure of the UBL domain of Dsk2 from S. cerevisiae | | Descriptor: | FORMIC ACID, UBIQUITIN-LIKE PROTEIN DSK2 | | Authors: | Lowe, E.D, Hasan, N, Trempe, J.-F, Fonso, L, Noble, M.E.M, Endicott, J.A, Johnson, L.N, Brown, N.R. | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structures of the Dsk2 Ubl and Uba Domains and Their Complex.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BWB
 
 | | Crystal structure of the UBA domain of Dsk2 from S. cerevisiae | | Descriptor: | UBIQUITIN-LIKE PROTEIN DSK2 | | Authors: | Lowe, E.D, Hasan, N, Trempe, J.-F, Fonso, L, Noble, M.E.M, Endicott, J.A, Johnson, L.N, Brown, N.R. | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Dsk2 Ubl and Uba Domains and Their Complex.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BWE
 
 | | The crystal structure of the complex between the UBA and UBL domains of Dsk2 | | Descriptor: | DSK2 | | Authors: | Lowe, E.D, Hasan, N, Trempe, J.-F, Fonso, L, Noble, M.E.M, Endicott, J.A, Johnson, L.N, Brown, N.R. | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the Dsk2 Ubl and Uba Domains and Their Complex.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2CCH
 
 | | The crystal structure of CDK2 cyclin A in complex with a substrate peptide derived from CDC modified with a gamma-linked ATP analogue | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION CONTROL PROTEIN 6 HOMOLOG, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Role of the Phospho-Cdk2/Cyclin a Recruitment Site in Substrate Recognition
J.Biol.Chem., 281, 2006
|
|
2CCI
 
 | | Crystal structure of phospho-CDK2 Cyclin A in complex with a peptide containing both the substrate and recruitment sites of CDC6 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6 homolog, Cyclin-A2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition.
J. Biol. Chem., 281, 2006
|
|
2ECP
 
 | | THE CRYSTAL STRUCTURE OF THE E. COLI MALTODEXTRIN PHOSPHORYLASE COMPLEX | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLYCEROL, MALTODEXTRIN PHOSPHORYLASE, ... | | Authors: | O'Reilly, M, Watson, K.A, Johnson, L.N. | | Deposit date: | 1998-10-27 | | Release date: | 1999-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of the Escherichia coli maltodextrin phosphorylase-acarbose complex.
Biochemistry, 38, 1999
|
|
5LYT
 
 | |
5LYZ
 
 | |
8LYZ
 
 | |
3LYZ
 
 | |
6LYT
 
 | |
