1T7N
 
 | |
5L1P
 
 | | X-ray Structure of Cytochrome P450 PntM with Pentalenolactone | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase, pentalenolactone | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1Q
 
 | | X-ray Structure of Cytochrome P450 PntM with Dihydropentalenolactone F | | Descriptor: | Dihydropentalenolactone F, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1O
 
 | | X-ray Structure of Cytochrome P450 PntM with Pentalenolactone F | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase, pentalenolactone F | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1W
 
 | |
5L1S
 
 | |
5L1V
 
 | |
5L1R
 
 | | X-ray Structure of the Substrate-free Cytochrome P450 PntM | | Descriptor: | BICINE, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1U
 
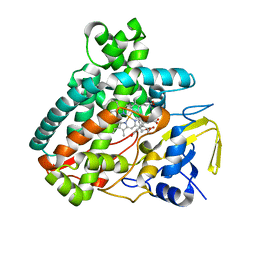 | |
5L1T
 
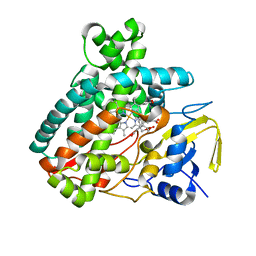 | |
2NXC
 
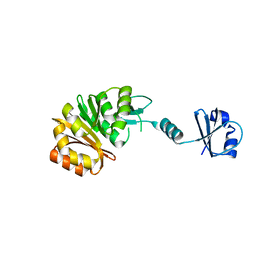 | |
3CJT
 
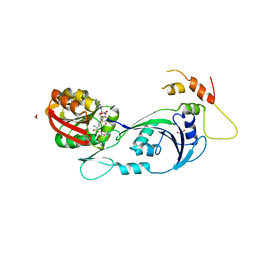 | | Ribosomal protein L11 methyltransferase (PrmA) in complex with dimethylated ribosomal protein L11 | | Descriptor: | 1,2-ETHANEDIOL, 50S ribosomal protein L11, CHLORIDE ION, ... | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-03-13 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple-Site Trimethylation of Ribosomal Protein L11 by the PrmA Methyltransferase.
Structure, 16, 2008
|
|
3CJR
 
 | | Ribosomal protein L11 methyltransferase (PrmA) in complex with ribosomal protein L11 (K39A) and inhibitor Sinefungin. | | Descriptor: | 50S ribosomal protein L11, Ribosomal protein L11 methyltransferase, SINEFUNGIN | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-03-13 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Multiple-Site Trimethylation of Ribosomal Protein L11 by the PrmA Methyltransferase.
Structure, 16, 2008
|
|
2NXJ
 
 | |
2NXN
 
 | | T. thermophilus ribosomal protein L11 methyltransferase (PrmA) in complex with ribosomal protein L11 | | Descriptor: | 50S ribosomal protein L11, Ribosomal protein L11 methyltransferase | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2006-11-17 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of ribosomal protein L11 by the protein trimethyltransferase PrmA.
Embo J., 26, 2007
|
|
2NXE
 
 | | T. thermophilus ribosomal protein L11 methyltransferase (PrmA) in complex with S-Adenosyl-L-Methionine | | Descriptor: | Ribosomal protein L11 methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2006-11-17 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recognition of ribosomal protein L11 by the protein trimethyltransferase PrmA.
Embo J., 26, 2007
|
|
3CJS
 
 | | Minimal Recognition Complex between PrmA and Ribosomal Protein L11 | | Descriptor: | 1,2-ETHANEDIOL, 50S ribosomal protein L11, Ribosomal protein L11 methyltransferase | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-03-13 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Multiple-Site Trimethylation of Ribosomal Protein L11 by the PrmA Methyltransferase.
Structure, 16, 2008
|
|
2OSV
 
 | | Crystal Structure of ZnuA from E. coli | | Descriptor: | High-affinity zinc uptake system protein znuA, ZINC ION | | Authors: | Li, H, Jogl, G. | | Deposit date: | 2007-02-06 | | Release date: | 2007-04-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Zinc-binding Transport Protein ZnuA from Escherichia coli Reveals an Unexpected Variation in Metal Coordination.
J.Mol.Biol., 368, 2007
|
|
3CJQ
 
 | | Ribosomal protein L11 methyltransferase (PrmA) in complex with dimethylated ribosomal protein L11 in space group P212121 | | Descriptor: | 50S ribosomal protein L11, IODIDE ION, N,N-dimethyl-L-methionine, ... | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-03-13 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Multiple-Site Trimethylation of Ribosomal Protein L11 by the PrmA Methyltransferase.
Structure, 16, 2008
|
|
3DMG
 
 | | T. Thermophilus 16S rRNA N2 G1207 methyltransferase (RsmC) in complex with AdoHcy | | Descriptor: | Probable ribosomal RNA small subunit methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-07-01 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Thermus thermophilus 16 S rRNA Methyltransferase RsmC in Complex with Cofactor and Substrate Guanosine.
J.Biol.Chem., 283, 2008
|
|
3DMH
 
 | | T. Thermophilus 16S rRNA N2 G1207 methyltransferase (RsmC) in complex with AdoMet and Guanosine | | Descriptor: | GUANOSINE, Probable ribosomal RNA small subunit methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-07-01 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Thermus thermophilus 16 S rRNA Methyltransferase RsmC in Complex with Cofactor and Substrate Guanosine.
J.Biol.Chem., 283, 2008
|
|
3DMF
 
 | | T. Thermophilus 16S rRNA N2 G1207 methyltransferase (RsmC) in complex with AdoMet | | Descriptor: | Probable ribosomal RNA small subunit methyltransferase, S-ADENOSYLMETHIONINE, SULFATE ION | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-07-01 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of the Thermus thermophilus 16 S rRNA Methyltransferase RsmC in Complex with Cofactor and Substrate Guanosine.
J.Biol.Chem., 283, 2008
|
|
3E9C
 
 | |
3EGV
 
 | | Ribosomal protein L11 methyltransferase (PrmA) in complex with trimethylated ribosomal protein L11 | | Descriptor: | 50S ribosomal protein L11, CHLORIDE ION, GLYCEROL, ... | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-09-11 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Multiple-site trimethylation of ribosomal protein L11 by the PrmA methyltransferase.
Structure, 16, 2008
|
|
3E9D
 
 | | Structure of full-length TIGAR from Danio rerio | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Zgc:56074 | | Authors: | Li, H, Jogl, G. | | Deposit date: | 2008-08-21 | | Release date: | 2008-12-16 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TIGAR (TP53 induced glycolysis and apoptosis regulator) is a fructose-2,6- and fructose-1,6-bisphosphatase
To be Published
|
|
