1EG2
 
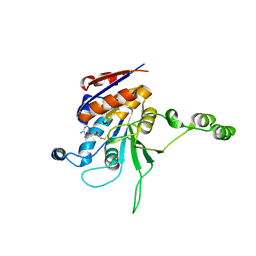 | | CRYSTAL STRUCTURE OF RHODOBACTER SPHEROIDES (N6 ADENOSINE) METHYLTRANSFERASE (M.RSRI) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MODIFICATION METHYLASE RSRI | | Authors: | Scavetta, R.D, Thomas, C.B, Walsh, M.A, Szegedi, S, Joachimiak, A, Gumport, R.I, Churchill, M.E.A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-02-11 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases.
Nucleic Acids Res., 28, 2000
|
|
2RC3
 
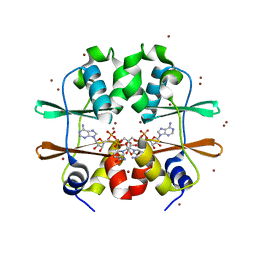 | | Crystal structure of CBS domain, NE2398 | | Descriptor: | BROMIDE ION, CBS domain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dong, A, Xu, X, Korniyenko, Y, Yakunin, A, Zheng, H, Walker, J.R, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-19 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of CBS domain, NE2398.
To be Published
|
|
1DPN
 
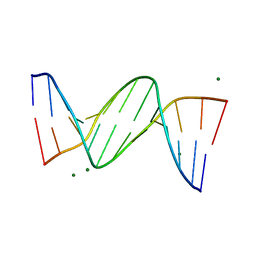 | | B-DODECAMER CGCGAA(TAF)TCGCG WITH INCORPORATED 2'-DEOXY-2'-FLUORO-ARABINO-FURANOSYL THYMINE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(TAF)P*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Egli, M, Tereshko, V, Teplova, M, Minasov, G, Joachimiak, A, Sanishvili, R, Weeks, C.M, Miller, R, Maier, M.A, An, H, Dan Cook, P, Manoharan, M. | | Deposit date: | 1999-12-27 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution.
Biopolymers, 48, 1998
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | Authors: | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X3Z
 
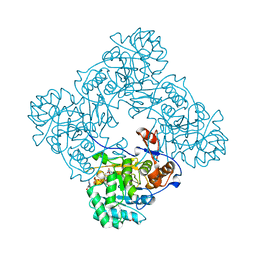 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP and NAD | | Descriptor: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Osipiuk, J, MALTSEVA, N, KIM, Y, Mulligan, R, MAKOWSKA-GRZYSKA, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP and NAD
to be published
|
|
4X2R
 
 | | Crystal structure of PriA from Actinomyces urogenitalis | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, PHOSPHATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-26 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Evolution of substrate specificity in a retained enzyme driven by gene loss.
Elife, 6, 2017
|
|
6AQE
 
 | | Crystal structure of PPK2 in complex with Mg ATP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nocek, B, Joachimiak, A, Yakunin, A. | | Deposit date: | 2017-08-19 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural Insights into Substrate Selectivity and Activity of Bacterial Polyphosphate Kinases
Acs Catalysis, 8, 2018
|
|
4X9S
 
 | | CRYSTAL STRUCTURE OF HISAP FROM STREPTOMYCES SP. MG1 | | Descriptor: | Phosphoribosyl isomerase A, SULFATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
4XEA
 
 | | Crystal structure of putative M16-like peptidase from Alicyclobacillus acidocaldarius | | Descriptor: | ACETATE ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Michalska, K, Tesar, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-23 | | Release date: | 2015-03-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of putative M16-like peptidase from Alicyclobacillus acidocaldarius
To Be Published
|
|
4XR9
 
 | | Crystal structure of CalS8 from Micromonospora echinospora cocrystallized with NAD and TDP-glucose | | Descriptor: | CalS8, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of CalS8 from Micromonospora echinospora
To Be Published
|
|
6ANH
 
 | |
6ANG
 
 | |
6AU0
 
 | | Crystal structure of PPK2 (Class III) in complex with bisphosphonate inhibitor (2-((3,5-dichlorophenyl)amino)ethane-1,1-diyl)diphosphonic acid | | Descriptor: | GLYCEROL, Polyphosphate:AMP phosphotransferase, {[(3,5-dichlorophenyl)amino]methylene}bis(phosphonic acid) | | Authors: | Nocek, B, Ruszkowski, M, Joachimiak, A, Berlicki, L, Yakunin, A. | | Deposit date: | 2017-08-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into Substrate Selectivity and Activity of Bacterial Polyphosphate Kinases
Acs Catalysis, 8, 2018
|
|
4XLT
 
 | | Crystal structure of response regulator receiver protein from Dyadobacter fermentans DSM 18053 | | Descriptor: | Response regulator receiver protein | | Authors: | Chang, C, Cuff, M, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-13 | | Release date: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of response regulator receiver protein from Dyadobacter fermentans DSM 18053
To Be Published
|
|
6AN9
 
 | |
1DNZ
 
 | | A-DNA DECAMER ACCGGCCGGT WITH MAGNESIUM BINDING SITES | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*CP*GP*GP*T)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-12-17 | | Release date: | 2000-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes.
Nucleic Acids Res., 28, 2000
|
|
5UPT
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis in complex with SBNP468 ligand | | Descriptor: | (7alpha,8alpha,10alpha,13alpha)-7,16-dihydroxykauran-18-oic acid, Acyl-CoA synthetase PtmA2, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Chang, C.Y, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
5UQP
 
 | | The crystal structure of cupin protein from Rhodococcus jostii RHA1 | | Descriptor: | CHLORIDE ION, Cupin, SULFATE ION, ... | | Authors: | Tan, K, Li, H, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-02-08 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of cupin protein from Rhodococcus jostii RHA1
To Be Published
|
|
5UVE
 
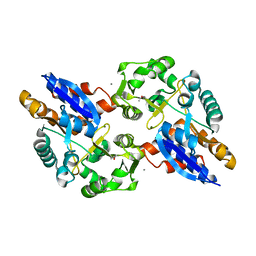 | | Crystal Structure of the ABC Transporter Substrate-binding protein BAB1_0226 from Brucella abortus | | Descriptor: | CALCIUM ION, GLYCEROL, Substrate-binding region of ABC-type glycine betaine transport system | | Authors: | Kim, Y, Chhor, G, Endres, M, Hero, J, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-02-20 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Beta-barrel-like Protein of Unknown Function
To Be Published
|
|
1DNT
 
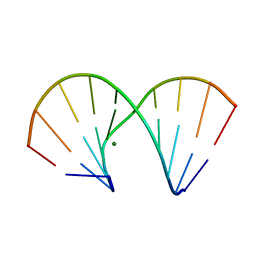 | | RNA/DNA DODECAMER R(GC)D(GTATACGC) WITH MAGNESIUM BINDING SITES | | Descriptor: | DNA/RNA (5'-R(*GP*CP)-D(*GP*TP*AP*TP*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-12-16 | | Release date: | 2000-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes.
Nucleic Acids Res., 28, 2000
|
|
5UZS
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P200 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-{[(4-chlorophenyl)carbamoyl]amino}propan-2-yl)-N-hydroxybenzene-1-carboximidamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-22 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.367 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P200
To Be Published
|
|
1DNX
 
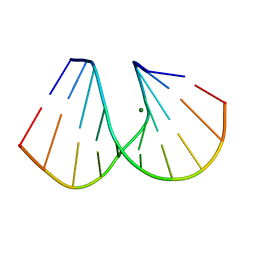 | | RNA/DNA DODECAMER R(G)D(CGTATACGC) WITH MAGNESIUM BINDING SITES | | Descriptor: | DNA/RNA (5'-R(*GP)-D(*CP*GP*TP*AP*TP*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-12-16 | | Release date: | 2000-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes.
Nucleic Acids Res., 28, 2000
|
|
5UUS
 
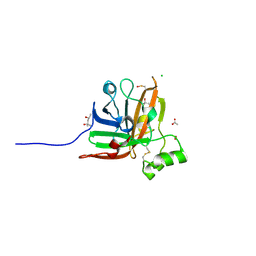 | | SrtF sortase from Corynebacterium diphtheriae | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Osipiuk, J, Gornicki, P, Ton-That, H, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-17 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | SrtF sortase from Corynebacterium diphtheriae
to be published
|
|
1DPL
 
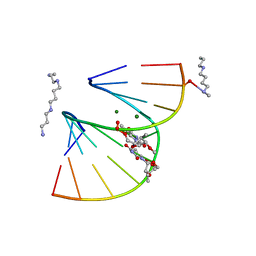 | | A-DNA DECAMER GCGTA(T23)TACGC WITH INCORPORATED 2'-METHOXY-3'-METHYLENEPHOSPHATE-THYMIDINE | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(T23)P*AP*CP*GP*C)-3', MAGNESIUM ION, SPERMINE | | Authors: | Egli, M, Tereshko, V, Teplova, M, Minasov, G, Joachimiak, A, Sanishvili, R, Weeks, C.M, Miller, R, Maier, M.A, An, H, Dan Cook, P, Manoharan, M. | | Deposit date: | 1999-12-27 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution.
Biopolymers, 48, 1998
|
|
5VES
 
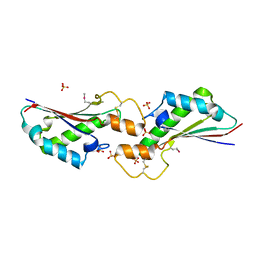 | | The 2.4A crystal structure of OmpA domain of OmpA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | Descriptor: | Outer membrane protein A, SULFATE ION | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2017-04-05 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into PG-binding, conformational change, and dimerization of the OmpA C-terminal domains from Salmonella enterica serovar Typhimurium and Borrelia burgdorferi.
Protein Sci., 26, 2017
|
|
