2KSS
 
 | | NMR structure of Myxococcus xanthus antirepressor CarS1 | | Descriptor: | Carotenogenesis protein carS | | Authors: | Jimenez, M, Gonzalez, C, Padmanabhan, S, Leon, E, Navarro-Aviles, G, Elias-Arnanz, M. | | Deposit date: | 2010-01-13 | | Release date: | 2010-05-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A bacterial antirepressor with SH3 domain topology mimics operator DNA in sequestering the repressor DNA recognition helix.
Nucleic Acids Res., 38, 2010
|
|
5T7M
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-TRP | | Descriptor: | ACETATE ION, Chemotaxis protein, SODIUM ION, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-05 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5LTO
 
 | | Ligand binding domain of Pseudomonas aeruginosa PAO1 amino acid chemoreceptors PctB in complex with L-Gln | | Descriptor: | GLUTAMINE, GLYCEROL, Methyl-accepting chemotaxis protein PctB, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.459 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
7QEK
 
 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-piruvic acid (IPA). | | Descriptor: | 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, MAGNESIUM ION, regulator AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
7QEJ
 
 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-acetic acid (IAA). | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MAGNESIUM ION, TRANSCRIPTIONAL REGULATOR AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
5T65
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-ILE | | Descriptor: | ACETATE ION, ISOLEUCINE, Methyl-accepting chemotaxis protein PctA, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-01 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5LT9
 
 | | Ligand binding domain of Pseudomonas aeruginosa PAO1 amino acid chemoreceptors PctB in complex with L-Arg | | Descriptor: | ARGININE, GLYCEROL, Methyl-accepting chemotaxis protein PctB, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
2C5R
 
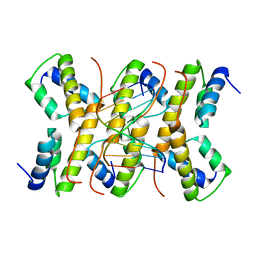 | | The structure of phage phi29 replication organizer protein p16.7 in complex with double stranded DNA | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*GP*GP*AP)-3', 5'-D(*TP*CP*CP*AP*CP*CP*GP*GP)-3', EARLY PROTEIN P16.7 | | Authors: | Albert, A, Jimenez, M, Munoz-Espin, D, Asensio, J.L, Hermoso, J.A, Salas, M, Meijer, W.J.J. | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Membrane Anchorage of Viral Phi 29 DNA During Replication.
J.Biol.Chem., 280, 2005
|
|
2IZ5
 
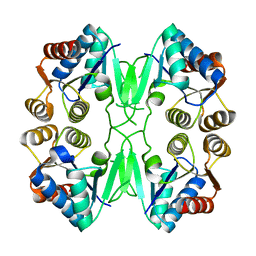 | | FUNCTION AND STRUCTURE OF THE MOLYBDENUM COFACTOR CARRIER PROTEIN MCP FROM CHLAMYDOMONAS REINHARDTII | | Descriptor: | MOCO CARRIER PROTEIN | | Authors: | Fischer, K, Llamas, A, Tejada-Jimenez, M, Schrader, N, Kuper, J, Mendel, R.R, Fernandez, E, Schwarz, G. | | Deposit date: | 2006-07-25 | | Release date: | 2006-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Function and Structure of the Molybdenum Cofactor Carrier Protein from Chlamydomonas Reinhardtii.
J.Biol.Chem., 281, 2006
|
|
2IZ7
 
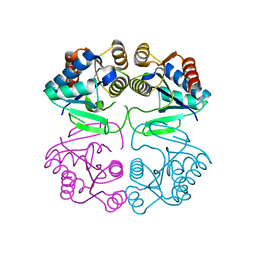 | | structure of Moco Carrier Protein from Chlamydomonas reinhardtii | | Descriptor: | MOCO CARRIER PROTEIN | | Authors: | Fischer, K, Llamas, A, Tejada-Jimenez, M, Schrader, N, Kuper, J, Mendel, R.R, Fernandez, E, Schwarz, G. | | Deposit date: | 2006-07-25 | | Release date: | 2006-07-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Function and Structure of the Molybdenum Cofactor Carrier Protein from Chlamydomonas Reinhardtii.
J.Biol.Chem., 281, 2006
|
|
2IZ6
 
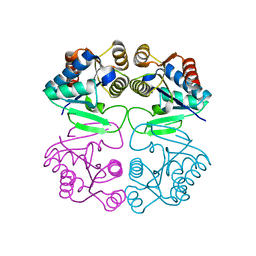 | | Structure of the Chlamydomonas rheinhardtii Moco Carrier Protein | | Descriptor: | MOLYBDENUM COFACTOR CARRIER PROTEIN | | Authors: | Fischer, K, Llamas, A, Tejada-Jimenez, M, Schrader, N, Kuper, J, Mendel, R.R, Fernandez, E, Schwarz, G. | | Deposit date: | 2006-07-25 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Function and Structure of the Molybdenum Cofactor Carrier Protein from Chlamydomonas Reinhardtii.
J.Biol.Chem., 281, 2006
|
|
2LWJ
 
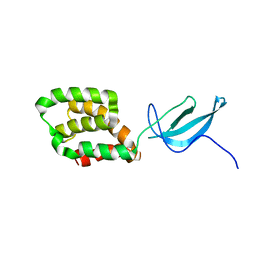 | |
2JML
 
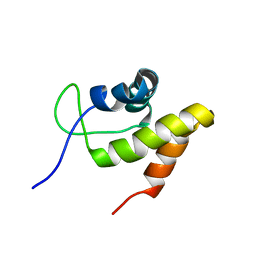 | | Solution structure of the N-terminal domain of CarA repressor | | Descriptor: | DNA BINDING DOMAIN/TRANSCRIPTIONAL REGULATOR | | Authors: | Jimenez, M, Padmanabhan, S, Gonzalez, C, Perez-Marin, M.C, Elias-Arnanz, M, Murillo, F.J, Rico, M. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for operator and antirepressor recognition by Myxococcus xanthus CarA repressor.
Mol.Microbiol., 63, 2007
|
|
2JOI
 
 | | NMR solution structure of hypothetical protein TA0095 from Thermoplasma acidophilum | | Descriptor: | Hypothetical protein Ta0095 | | Authors: | Jimenez, M, Leon, E, Santoro, J, Rico, M, Yee, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-13 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical protein TA0095 from Thermoplasma acidophilum: A novel superfamily with a two-layer sandwich architecture
Protein Sci., 16, 2007
|
|
2NCT
 
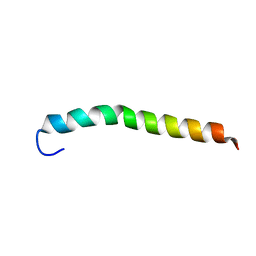 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
2NCS
 
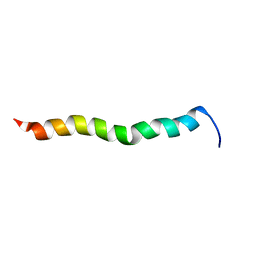 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
2LQK
 
 | |
2MWT
 
 | | NMR structure of crotalicidin in DPC micelles | | Descriptor: | Cathelicidin-like peptide | | Authors: | Jimenez, M, Zamora-Carreras, H. | | Deposit date: | 2014-11-24 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Dissection of Crotalicidin, a Rattlesnake Venom Cathelicidin, Retrieves a Fragment with Antimicrobial and Antitumor Activity.
J.Med.Chem., 58, 2015
|
|
1RML
 
 | | NMR STUDY OF ACID FIBROBLAST GROWTH FACTOR BOUND TO 1,3,6-NAPHTHALENE TRISULPHONATE, 26 STRUCTURES | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR, NAPHTHALENE TRISULFONATE | | Authors: | Lozano, R.M, Jimenez, M.A, Santoro, J, Rico, M, Gimenez-Gallego, G. | | Deposit date: | 1998-05-21 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of acidic fibroblast growth factor bound to 1,3, 6-naphthalenetrisulfonate: a minimal model for the anti-tumoral action of suramins and suradistas.
J.Mol.Biol., 281, 1998
|
|
2LT4
 
 | |
5LTV
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECPETOR PCTC IN COMPLEX WITH GABA | | Descriptor: | ACETATE ION, Chemotactic transducer PctC, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
2JW4
 
 | | NMR solution structure of the N-terminal SH3 domain of human Nckalpha | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Santiveri, C.M, Borroto, A, Simon, L, Rico, M, Ortiz, A.R, Alarcon, B, Jimenez, M. | | Deposit date: | 2007-10-05 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Interaction between the N-terminal SH3 domain of Nckalpha and CD3epsilon-derived peptides: Non-canonical and canonical recognition motifs
BIOCHEM.BIOPHYS.ACTA PROTEINS & PROTEOMICS, 1794, 2009
|
|
2KB5
 
 | | Solution NMR Structure of Eosinophil Cationic Protein/RNase 3 | | Descriptor: | Eosinophil cationic protein | | Authors: | Rico, M, Bruix, M, Laurents, D.V, Santoro, J, Jimenez, M, Boix, E, Moussaoui, M, Nogues, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-06-23 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The (1)H, (13)C, (15)N resonance assignment, solution structure, and residue level stability of eosinophil cationic protein/RNase 3 determined by NMR spectroscopy
Biopolymers, 91, 2009
|
|
6YMZ
 
 | | Structure of the CheB methylsterase from P. atrosepticum SCRI1043 | | Descriptor: | ACETATE ION, GLYCEROL, Protein-glutamate methylesterase/protein-glutamine glutaminase, ... | | Authors: | Gavira, J.A, Krell, T, Velando-Soriano, F, Matilla, M.A. | | Deposit date: | 2020-04-10 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence for Pentapeptide-Dependent and Independent CheB Methylesterases.
Int J Mol Sci, 21, 2020
|
|
2M8O
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in DPC | | Descriptor: | Transmembrane protein gp41 | | Authors: | Serrano, S, Huarte, N, Nieva, J.L, Jimenez, M. | | Deposit date: | 2013-05-23 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Immunogenicity of a Peptide Vaccine, Including the Complete HIV-1 gp41 2F5 Epitope: IMPLICATIONS FOR ANTIBODY RECOGNITION MECHANISM AND IMMUNOGEN DESIGN.
J.Biol.Chem., 289, 2014
|
|
