2AXW
 
 | | Structure of DraD invasin from uropathogenic Escherichia coli | | 分子名称: | CHLORIDE ION, DraD invasin, GLYCEROL | | 著者 | Jedrzejczak, R, Dauter, Z, Dauter, M, Piatek, R, Zalewska, B, Mroz, M, Bury, K, Nowicki, B, Kur, J. | | 登録日 | 2005-09-06 | | 公開日 | 2005-11-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Structure of DraD invasin from uropathogenic Escherichia coli: a dimer with swapped beta-tails.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2FXQ
 
 | |
3RC3
 
 | | Human Mitochondrial Helicase Suv3 | | 分子名称: | ATP-dependent RNA helicase SUPV3L1, mitochondrial, AZIDE ION, ... | | 著者 | Dauter, Z, Jedrzejczak, R, Dauter, M, Szczesny, R, Stepien, P. | | 登録日 | 2011-03-30 | | 公開日 | 2011-05-11 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Human Suv3 protein reveals unique features among SF2 helicases.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3RC8
 
 | | Human Mitochondrial Helicase Suv3 in Complex with Short RNA Fragment | | 分子名称: | ATP-dependent RNA helicase SUPV3L1, mitochondrial, RNA fragment | | 著者 | Dauter, Z, Jedrzejczak, R, Dauter, M, Wang, J, Szczesny, R, Stepien, P. | | 登録日 | 2011-03-30 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Human Suv3 protein reveals unique features among SF2 helicases.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4WHI
 
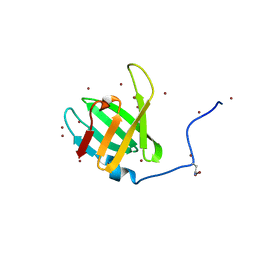 | | Crystal structure of C-terminal domain of penicillin binding protein Rv0907 | | 分子名称: | BROMIDE ION, Beta-lactamase, NICKEL (II) ION | | 著者 | Chang, C, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-09-22 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of C-terminal domain of penicillin binding protein Rv0907
To Be Published
|
|
7N3C
 
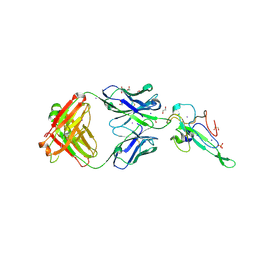 | | Crystal Structure of Human Fab S24-202 in the complex with the N-terminal Domain of Nucleocapsid protein from SARS CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, IODIDE ION, Nucleoprotein, ... | | 著者 | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-05-31 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7N3D
 
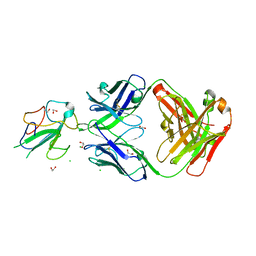 | | Crystal Structure of Human Fab S24-1564 in the complex with the N-terminal Domain of Nucleocapsid protein from SARS CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Nucleoprotein, ... | | 著者 | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-05-31 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7THH
 
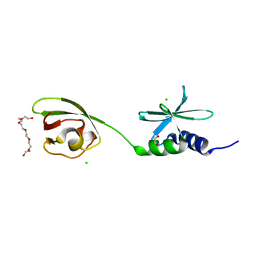 | | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | 著者 | Osipiuk, J, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-11 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein
to be published
|
|
6XKM
 
 | | Room Temperature Structure of SARS-CoV-2 NSP10/NSP16 Methyltransferase in a Complex with SAM Determined by Fixed-Target Serial Crystallography | | 分子名称: | 2'-O-methyltransferase, CHLORIDE ION, Non-structural protein 10, ... | | 著者 | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-26 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | 分子名称: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2023-02-14 | | 公開日 | 2023-02-22 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
8GHY
 
 | | Crystal Structure of the E154D mutant CelD Cellulase from the Anaerobic Fungus Piromyces finnis in the complex with cellotriose. | | 分子名称: | Cellulase CelD, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Dementieve, A, Kim, Y, Jedrzejczak, R, Michalska, K, Joachimiak, A. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
8GHX
 
 | | Crystal Structure of CelD Cellulase from the Anaerobic Fungus Piromyces finnis | | 分子名称: | 1,2-ETHANEDIOL, Cellulase CelD | | 著者 | Dementieve, A, Kim, Y, Jedrzejczak, R, Michalska, K, Joachimiak, A. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
6XIP
 
 | | The 1.5 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | 著者 | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-20 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
5IR2
 
 | | Crystal structure of novel cellulases from microbes associated with the gut ecosystem | | 分子名称: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Cellulase, ... | | 著者 | Chang, C, Mack, J, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2016-03-11 | | 公開日 | 2016-03-23 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.079 Å) | | 主引用文献 | Crystal structure of novel cellulases from microbes associated with the gut ecosystem
To Be Published
|
|
5JBR
 
 | | Crystal structure of uncharacterized protein Bcav_2135 from Beutenbergia cavernae | | 分子名称: | SULFATE ION, Uncharacterized protein Bcav_2135 | | 著者 | Chang, C, Cuff, M, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2016-04-13 | | 公開日 | 2016-04-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of uncharacterized protein Bcav_2135 from Beutenbergia cavernae
To Be Published
|
|
5JMB
 
 | |
4YF1
 
 | | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e | | 分子名称: | CITRATE ANION, Lmo0812 protein, SODIUM ION | | 著者 | Krishna, S.N, Light, S.H, Filippova, E.V, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-02-24 | | 公開日 | 2015-03-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e
To Be Published
|
|
4ZR7
 
 | | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168 | | 分子名称: | ACETATE ION, CHLORIDE ION, Sensor histidine kinase ResE | | 著者 | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-11 | | 公開日 | 2015-05-27 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
4ZNM
 
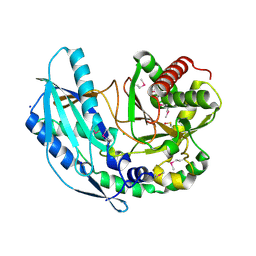 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form) | | 分子名称: | C-domain type II peptide synthetase, CHLORIDE ION, SODIUM ION | | 著者 | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-05-04 | | 公開日 | 2015-05-27 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form)
To Be Published
|
|
4ZXW
 
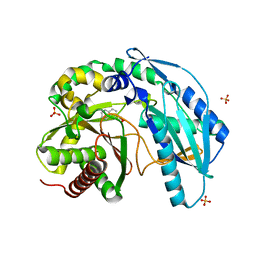 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (complex with (R)-(-)-1-(2-naphthyl)-1,2-ethanediol and sucrose) | | 分子名称: | (1R)-1-(naphthalen-2-yl)ethane-1,2-diol, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-domain type II peptide synthetase, ... | | 著者 | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-05-20 | | 公開日 | 2015-06-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.187 Å) | | 主引用文献 | Crystal structure of SgcC5 protein from Streptomyces globisporus
To Be Published
|
|
5CQF
 
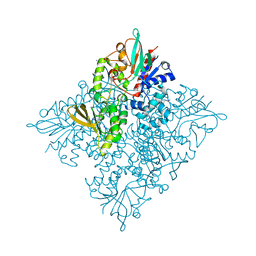 | | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae | | 分子名称: | IODIDE ION, L-lysine 6-monooxygenase | | 著者 | Michalska, K, Bigelow, L, Jedrzejczak, R, Weerth, R.S, Cao, H, Yennamalli, R, Phillips Jr, G.N, Thomas, M.G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-07-21 | | 公開日 | 2015-09-30 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae
To Be Published
|
|
5F4Z
 
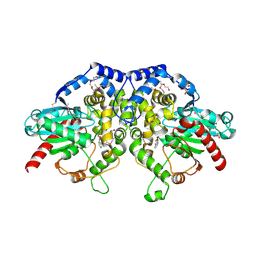 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus | | 分子名称: | (1~{R},2~{R})-2,3-dihydro-1~{H}-indene-1,2-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | 著者 | Tan, K, Li, H, Jedrzejczak, R, BABNIGG, G, BINGMAN, C.A, YENNAMALLI, R, LOHMAN, J, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-12-03 | | 公開日 | 2016-02-17 | | 最終更新日 | 2020-09-23 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus
To Be Published
|
|
6AZY
 
 | | Crystal structure of Hsp104 R328M/R757M mutant from Calcarisporiella thermophila | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein Hsp104 | | 著者 | Michalska, K, Bigelow, L, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-09-13 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
6C9Z
 
 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) W169Y mutant FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose | | 分子名称: | (1S,2S,3R,4S,5S)-5-[(1,3-dihydroxypropan-2-yl)amino]-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, Glycosyl hydrolase, family 31 | | 著者 | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | 登録日 | 2018-01-29 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) W169Y mutant FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose
To Be Published
|
|
6CA3
 
 | | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol | | 分子名称: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, GLYCEROL, Glycosyl hydrolase, ... | | 著者 | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | 登録日 | 2018-01-29 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.743 Å) | | 主引用文献 | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol
To Be Published
|
|
