8PND
 
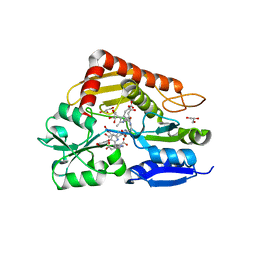 | | The ES3 intermediate of hydroxymethylbilane synthase R167Q variant | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, ... | | Authors: | Saeter, M.C, Bustad, H.J, Laitaoja, M, Janis, J, Martinez, A, Aarsand, A.K, Kallio, J.P. | | Deposit date: | 2023-06-30 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | One ring closer to a closure: the crystal structure of the ES 3 hydroxymethylbilane synthase intermediate.
Febs J., 291, 2024
|
|
1XNK
 
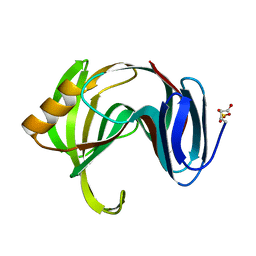 | | Beta-1,4-xylanase from Chaetomium thermophilum complexed with methyl thioxylopentoside | | Descriptor: | 4-thio-beta-D-xylopyranose-(1-4)-4-thio-beta-D-xylopyranose-(1-4)-methyl 4-thio-alpha-D-xylopyranoside, SULFATE ION, endoxylanase 11A | | Authors: | Hakanpaa, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2004-10-05 | | Release date: | 2005-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Determination of thioxylo-oligosaccharide binding to family 11 xylanases using electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry and X-ray crystallography
FEBS J., 272, 2005
|
|
4BCS
 
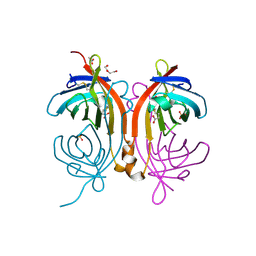 | | Crystal structure of an avidin mutant | | Descriptor: | ACETATE ION, BIOTIN, CHIMERIC AVIDIN, ... | | Authors: | Airenne, T.T, Niederhauser, B, Hytonen, V.P, Kulomaa, M.S, Johnson, M.S. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel Chimeric Avidin with Increased Thermal Stability Using DNA Shuffling.
Plos One, 9, 2014
|
|
4UR8
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with 2-oxoadipic acid | | Descriptor: | 2-OXOADIPIC ACID, FORMIC ACID, KETO-DEOXY-D-GALACTARATE DEHYDRATASE | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure and Function of a Decarboxylating Agrobacterium Tumefaciens Keto-Deoxy-D-Galactarate Dehydratase.
Biochemistry, 53, 2014
|
|
7AAJ
 
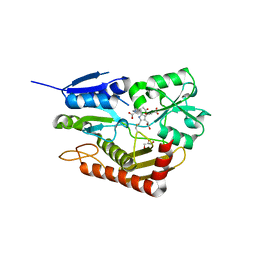 | | Human porphobilinogen deaminase in complex with cofactor | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, GLYCEROL, Porphobilinogen deaminase | | Authors: | Kallio, J.P, Bustad, H.J, Martinez, A. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of porphobilinogen deaminase mutants reveals that arginine-173 is crucial for polypyrrole elongation mechanism.
Iscience, 24, 2021
|
|
7AAK
 
 | | Human porphobilinogen deaminase R173W mutant crystallized in the ES2 intermediate state | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, Porphobilinogen deaminase | | Authors: | Kallio, J.P, Bustad, H.J, Martinez, A. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of porphobilinogen deaminase mutants reveals that arginine-173 is crucial for polypyrrole elongation mechanism.
Iscience, 24, 2021
|
|
7ZSC
 
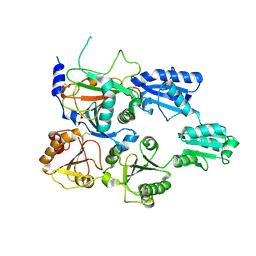 | | Crystal structure of the heterodimeric human C-P4H-II with truncated alpha subunit (C-P4H-II delta281) | | Descriptor: | Prolyl 4-hydroxylase subunit alpha-2, Protein disulfide-isomerase, SULFATE ION | | Authors: | Lebedev, A, Koski, M.K, Wierenga, R.K, Murthy, A.V, Sulu, R. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Crystal structure of the collagen prolyl 4-hydroxylase (C-P4H) catalytic domain complexed with PDI: Toward a model of the C-P4H alpha 2 beta 2 tetramer.
J.Biol.Chem., 298, 2022
|
|
5HWM
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with 2-oxoadipic acid | | Descriptor: | 2-OXOADIPIC ACID, FORMIC ACID, Probable 5-dehydro-4-deoxyglucarate dehydratase | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure and function of a decarboxylating Agrobacterium tumefaciens keto-deoxy-d-galactarate dehydratase.
Biochemistry, 53, 2014
|
|
1H1A
 
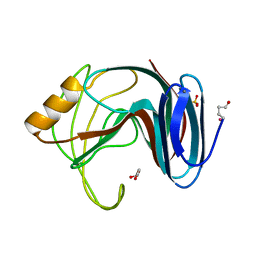 | | Thermophilic beta-1,4-xylanase from Chaetomium thermophilum | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, GLYCEROL, ... | | Authors: | Hakulinen, N, Rouvinen, J. | | Deposit date: | 2002-07-05 | | Release date: | 2003-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-Dimensional Structures of Thermophilic Beta-1,4-Xylanases from Chaetomium Thermophilum and Nonomuraea Flexuosa. Comparison of Twelve Xylanases in Relation to Their Thermal Stability.
Eur.J.Biochem., 270, 2003
|
|
4UR7
 
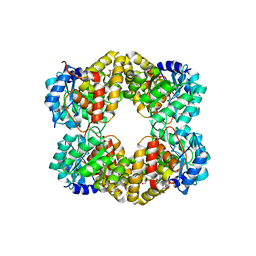 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with pyruvate | | Descriptor: | FORMIC ACID, GLYCEROL, KETO-DEOXY-D-GALACTARATE DEHYDRATASE | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure and Function of a Decarboxylating Agrobacterium Tumefaciens Keto-Deoxy-D-Galactarate Dehydratase.
Biochemistry, 53, 2014
|
|
4WFU
 
 | |
4WFV
 
 | |
3FU7
 
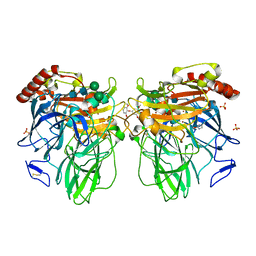 | | Melanocarpus albomyces laccase crystal soaked (4 sec) with 2,6-dimethoxyphenol | | Descriptor: | 2,6-dimethoxycyclohexa-2,5-diene-1,4-dione, 2,6-dimethoxyphenol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure Function Studies of a Melanocarpus albomyces Laccase Suggest a Pathway for Oxidation of Phenolic Compounds.
J.Mol.Biol., 392, 2009
|
|
2N1U
 
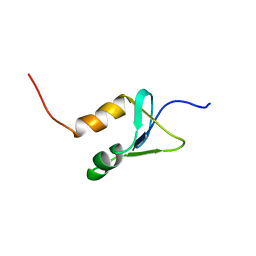 | | Structure of SAP30L corepressor protein | | Descriptor: | Histone deacetylase complex subunit SAP30L, ZINC ION | | Authors: | Tossavainen, H, Permi, P. | | Deposit date: | 2015-04-23 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Redox-dependent disulfide bond formation in SAP30L corepressor protein: Implications for structure and function.
Protein Sci., 25, 2016
|
|
8RTW
 
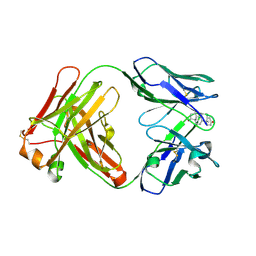 | | Crystal Structure of the Anti-testosterone Fab in Complex with Testosterone | | Descriptor: | Anti-testosterone Fab 220 heavy chain, Anti-testosterone Fab 220 light chain, TESTOSTERONE | | Authors: | Eronen, V, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into ternary immunocomplex formation and cross-reactivity: binding of an anti-immunocomplex FabB12 to Fab220-testosterone complex.
Febs J., 291, 2024
|
|
8RTX
 
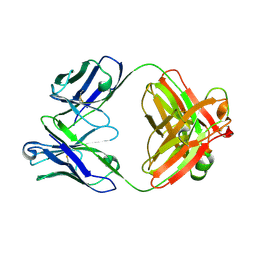 | | Crystal Structure of an Anti-idiotype Fab Fragment | | Descriptor: | Anti-IC Fab fragment B12 heavy chain, Anti-IC Fab fragment B12 light chain | | Authors: | Eronen, V, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into ternary immunocomplex formation and cross-reactivity: binding of an anti-immunocomplex FabB12 to Fab220-testosterone complex.
Febs J., 291, 2024
|
|
2JGS
 
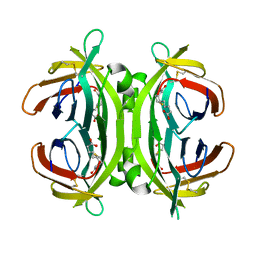 | | Circular permutant of avidin | | Descriptor: | BIOTIN, CIRCULAR PERMUTANT OF AVIDIN | | Authors: | Maatta, J.A.E, Hytonen, V.P, Airenne, T.T, Niskanen, E, Johnson, M.S, Kulomaa, M.S, Nordlund, H.R. | | Deposit date: | 2007-02-14 | | Release date: | 2008-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Modification of Ligand-Binding Preference of Avidin by Circular Permutation and Mutagenesis.
Chembiochem, 9, 2008
|
|
4ODD
 
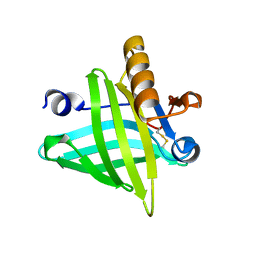 | |
4BJ8
 
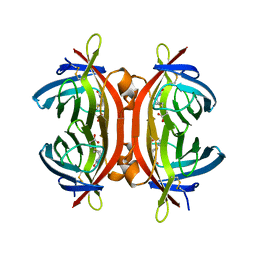 | | Zebavidin | | Descriptor: | BIOTIN, GLYCEROL, ZEBAVIDIN | | Authors: | Airenne, T.T, Parthiban, M, Niederhauser, B, Zmurko, J, Kulomaa, M.S, Hytonen, V.P, Johnson, M.S. | | Deposit date: | 2013-04-17 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Zebavidin
Plos One, 8, 2013
|
|
8QPS
 
 | |
3FU9
 
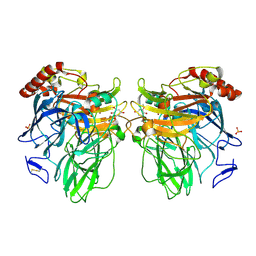 | | Melanocarpus albomyces laccase crystal soaked (20 min) with 2,6-dimethoxyphenol | | Descriptor: | 2,6-dimethoxybenzene-1,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-09-22 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Function Studies of a Melanocarpus albomyces Laccase Suggest a Pathway for Oxidation of Phenolic Compounds.
J.Mol.Biol., 392, 2009
|
|
3FU8
 
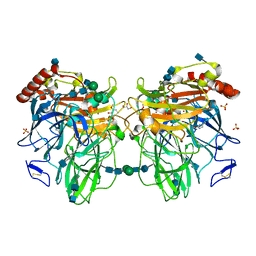 | | Melanocarpus albomyces laccase crystal soaked (10 sec) with 2,6-dimethoxyphenol | | Descriptor: | 2,6-dimethoxyphenol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-09-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Studies of a Melanocarpus albomyces Laccase Suggest a Pathway for Oxidation of Phenolic Compounds.
J.Mol.Biol., 392, 2009
|
|
4GGZ
 
 | | The structure of bradavidin2-biotin complex | | Descriptor: | BIOTIN, Bradavidin 2 | | Authors: | Livnah, O, Meir, A. | | Deposit date: | 2012-08-07 | | Release date: | 2013-06-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The highly dynamic oligomeric structure of bradavidin II is unique among avidin proteins.
Protein Sci., 22, 2013
|
|
4GGR
 
 | | The structure of apo bradavidin2 (Form A) | | Descriptor: | Bradavidin 2 | | Authors: | Livnah, O, Meir, A. | | Deposit date: | 2012-08-07 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The highly dynamic oligomeric structure of bradavidin II is unique among avidin proteins.
Protein Sci., 22, 2013
|
|
4GGT
 
 | | Structure of apo Bradavidin2 (Form B) | | Descriptor: | Bradavidin 2 | | Authors: | Livnah, O, Meir, A. | | Deposit date: | 2012-08-07 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | The highly dynamic oligomeric structure of bradavidin II is unique among avidin proteins.
Protein Sci., 22, 2013
|
|
