1AVF
 
 | | ACTIVATION INTERMEDIATE 2 OF HUMAN GASTRICSIN FROM HUMAN STOMACH | | Descriptor: | GASTRICSIN, SODIUM ION | | Authors: | Khan, A.R, Cherney, M.M, Tarasova, N.I, James, M.N.G. | | Deposit date: | 1997-09-16 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural characterization of activation 'intermediate 2' on the pathway to human gastricsin.
Nat.Struct.Biol., 4, 1997
|
|
1CT4
 
 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-VAL18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
1CSO
 
 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-ILE18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
1CT0
 
 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-SER18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
1CHO
 
 | | CRYSTAL AND MOLECULAR STRUCTURES OF THE COMPLEX OF ALPHA-*CHYMOTRYPSIN WITH ITS INHIBITOR TURKEY OVOMUCOID THIRD DOMAIN AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | ALPHA-CHYMOTRYPSIN A, TURKEY OVOMUCOID THIRD DOMAIN (OMTKY3) | | Authors: | Fujinaga, M, Sielecki, A.R, Read, R.J, Ardelt, W, Laskowskijunior, M, James, M.N.G. | | Deposit date: | 1988-03-04 | | Release date: | 1988-07-16 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and molecular structures of the complex of alpha-chymotrypsin with its inhibitor turkey ovomucoid third domain at 1.8 A resolution.
J.Mol.Biol., 195, 1987
|
|
2NQT
 
 | | Crystal structure of N-Acetyl-gamma-Glutamyl-Phosphate Reductase (Rv1652) from Mycobacterium tuberculosis at 1.58 A resolution | | Descriptor: | N-acetyl-gamma-glutamyl-phosphate reductase | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Moraidin, F, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of N-acetyl-gamma-glutamyl-phosphate Reductase from Mycobacterium tuberculosis in Complex with NADP(+).
J.Mol.Biol., 367, 2007
|
|
2H9H
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | Hepatitis A virus protease 3C, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Three residue peptide | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-06-09 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | An Episulfide Cation (Thiiranium Ring) Trapped in the Active Site of HAV 3C Proteinase Inactivated by Peptide-based Ketone Inhibitors.
J.Mol.Biol., 361, 2006
|
|
2Q9J
 
 | | Crystal structure of the C217S mutant of diaminopimelate epimerase | | Descriptor: | 1,2-ETHANEDIOL, Diaminopimelate epimerase, SULFATE ION | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2P2G
 
 | | Crystal Structure of Ornithine Carbamoyltransferase from Mycobacterium Tuberculosis (Rv1656): Orthorhombic Form | | Descriptor: | Ornithine carbamoyltransferase, SULFATE ION | | Authors: | Sankaranarayanan, R, Cherney, M.M, Cherney, L.T, Garen, C, Moradian, F, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-03-07 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structures of ornithine carbamoyltransferase from Mycobacterium tuberculosis and its ternary complex with carbamoyl phosphate and L-norvaline reveal the enzyme's catalytic mechanism.
J.Mol.Biol., 375, 2008
|
|
1ESB
 
 | | DIRECT STRUCTURE OBSERVATION OF AN ACYL-ENZYME INTERMEDIATE IN THE HYDROLYSIS OF AN ESTER SUBSTRATE BY ELASTASE | | Descriptor: | CALCIUM ION, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, PORCINE PANCREATIC ELASTASE, ... | | Authors: | Ding, X, Rasmussen, B, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-04 | | Release date: | 1994-04-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Direct structural observation of an acyl-enzyme intermediate in the hydrolysis of an ester substrate by elastase.
Biochemistry, 33, 1994
|
|
1ER8
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | Endothiapepsin, H-77 | | Authors: | Hemmings, A.M, Veerapandian, B, Szelke, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1989-10-16 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
2QAA
 
 | |
2QA9
 
 | |
2QVB
 
 | | Crystal Structure of Haloalkane Dehalogenase Rv2579 from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Haloalkane dehalogenase 3 | | Authors: | Mazumdar, P.A, Hulecki, J, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-08-08 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | X-ray crystal structure of Mycobacterium tuberculosis haloalkane dehalogenase Rv2579.
Biochim.Biophys.Acta, 1784, 2008
|
|
2REN
 
 | | STRUCTURE OF RECOMBINANT HUMAN RENIN, A TARGET FOR CARDIOVASCULAR-ACTIVE DRUGS, AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RENIN | | Authors: | Sielecki, A.R, James, M.N.G. | | Deposit date: | 1992-02-05 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of recombinant human renin, a target for cardiovascular-active drugs, at 2.5 A resolution.
Science, 243, 1989
|
|
2FMJ
 
 | |
2GKR
 
 | | Crystal structure of the N-terminally truncated OMTKY3-del(1-5) | | Descriptor: | CHLORIDE ION, Ovomucoid | | Authors: | Lee, T.W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-04-03 | | Release date: | 2007-02-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural Insights into the Non-additivity Effects in the Sequence-to-Reactivity Algorithm for Serine Peptidases and their Inhibitors.
J.Mol.Biol., 367, 2007
|
|
2GKV
 
 | | Crystal structure of the SGPB:P14'-Ala32 OMTKY3-del(1-5) complex | | Descriptor: | Ovomucoid, Streptogrisin B | | Authors: | Lee, T.W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-04-03 | | Release date: | 2007-02-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Non-additivity Effects in the Sequence-to-Reactivity Algorithm for Serine Peptidases and their Inhibitors.
J.Mol.Biol., 367, 2007
|
|
2GKT
 
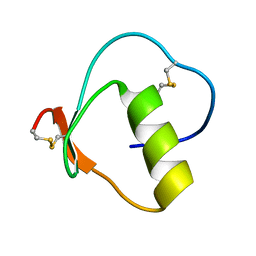 | |
2HRV
 
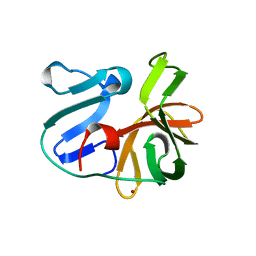 | | 2A CYSTEINE PROTEINASE FROM HUMAN RHINOVIRUS 2 | | Descriptor: | 2A CYSTEINE PROTEINASE, ZINC ION | | Authors: | Petersen, J.F.W, Cherney, M.M, Liebig, H.-D, Skern, T, Kuechler, E, James, M.N.G. | | Deposit date: | 1999-04-29 | | Release date: | 2000-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of the 2A proteinase from a common cold virus: a proteinase responsible for the shut-off of host-cell protein synthesis.
EMBO J., 18, 1999
|
|
1HAV
 
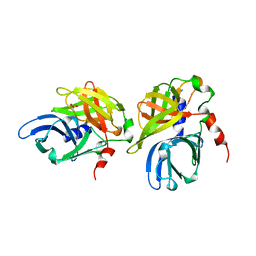 | | HEPATITIS A VIRUS 3C PROTEINASE | | Descriptor: | CHLORIDE ION, HEPATITIS A VIRUS 3C PROTEINASE | | Authors: | Bergmann, E.M, James, M.N.G. | | Deposit date: | 1996-10-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition.
J.Virol., 71, 1997
|
|
5ER1
 
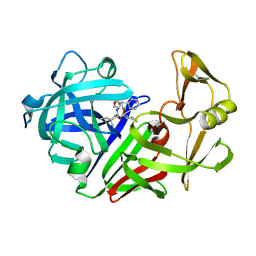 | |
1GBF
 
 | |
1GBD
 
 | |
1GBK
 
 | |
