1OUS
 
 | | Lecb (PA-LII) calcium-free | | Descriptor: | SULFATE ION, hypothetical protein LecB | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
1OVS
 
 | | LecB (PA-LII) in complex with core trimannoside | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose, ... | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-27 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
1OXC
 
 | | LecB (PA-LII) in complex with FUCOSE | | Descriptor: | CALCIUM ION, SULFATE ION, alpha-L-fucopyranose, ... | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-04-02 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
1OVP
 
 | | LecB (PA-LII) in complex with fructose | | Descriptor: | CALCIUM ION, beta-D-fructopyranose, hypothetical protein LecB | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-27 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
1OUX
 
 | | LecB (PA-LII) sugar-free | | Descriptor: | CALCIUM ION, SULFATE ION, hypothetical protein LecB | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
1OUR
 
 | | LecB (PA-LII) in complex with mannose | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose, hypothetical protein LecB | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
1I6W
 
 | | THE CRYSTAL STRUCTURE OF BACILLUS SUBTILIS LIPASE: A MINIMAL ALPHA/BETA HYDROLASE ENZYME | | Descriptor: | CADMIUM ION, LIPASE A | | Authors: | van Pouderoyen, G, Eggert, T, Jaeger, K.-E, Dijkstra, B.W. | | Deposit date: | 2001-03-05 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of Bacillus subtilis lipase: a minimal alpha/beta hydrolase fold enzyme.
J.Mol.Biol., 309, 2001
|
|
6Z68
 
 | | A novel metagenomic alpha/beta-fold esterase | | Descriptor: | Acetyl esterase/lipase, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Bollinger, A, Thies, S, Hoeppner, A, Kobus, S, Jaeger, K.-E, Smits, S.H.J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of a novel family IV esterase in free and substrate-bound form.
Febs J., 288, 2021
|
|
4JGG
 
 | |
4KUK
 
 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Dark state) | | Descriptor: | ACETIC ACID, RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
4KUO
 
 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Photoexcited state) | | Descriptor: | RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
5OVM
 
 | | Solution structure of lipase binding domain LID1 of foldase from Pseudomonas aeruginosa | | Descriptor: | Lipase chaperone | | Authors: | Viegas, A, Jaeger, K.-E, Etzkorn, M, Gohlke, H, Verma, N, Dollinger, P, Kovacic, F. | | Deposit date: | 2017-08-29 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic insights revealing how lipase binding domain MD1 of Pseudomonas aeruginosa foldase affects lipase activation.
Sci Rep, 10, 2020
|
|
6Z69
 
 | | A novel metagenomic alpha/beta-fold esterase | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Acetyl esterase/lipase, MAGNESIUM ION, ... | | Authors: | Bollinger, A, Thies, S, Hoeppner, A, Kobus, S, Jaeger, K.-E, Smits, S.H.J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of a novel family IV esterase in free and substrate-bound form.
Febs J., 288, 2021
|
|
6SCD
 
 | | Polyester hydrolase PE-H Y250S mutant of Pseudomonas aestusnigri | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bollinger, A, Thies, S, Kobus, S, Hoeppner, A, Smits, S.H.J, Jaeger, K.-E. | | Deposit date: | 2019-07-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Novel Polyester Hydrolase From the Marine BacteriumPseudomonas aestusnigri -Structural and Functional Insights.
Front Microbiol, 11, 2020
|
|
6SBN
 
 | | Polyester hydrolase PE-H of Pseudomonas aestusnigri | | Descriptor: | ACETATE ION, SODIUM ION, polyester hydrolase | | Authors: | Bollinger, A, Thies, S, Kobus, S, Hoeppner, A, Smits, S.H.J, Jaeger, K.-E. | | Deposit date: | 2019-07-22 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A Novel Polyester Hydrolase From the Marine BacteriumPseudomonas aestusnigri -Structural and Functional Insights.
Front Microbiol, 11, 2020
|
|
4FBM
 
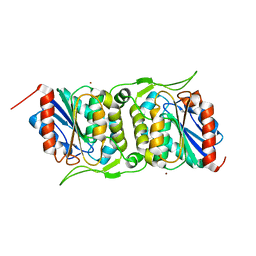 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | BROMIDE ION, LipS lipolytic enzyme | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovavic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
4FBL
 
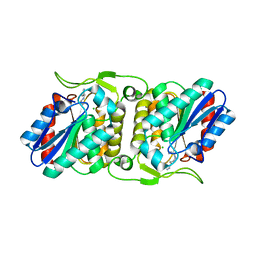 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | CHLORIDE ION, LipS lipolytic enzyme, SPERMIDINE | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovacic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
6GSF
 
 | | Solution structure of lipase binding domain LID1 of foldase from Pseudomonas aeruginosa | | Descriptor: | Lipase chaperone | | Authors: | Viegas, A, Jaeger, K.-E, Etzkorn, M, Gohlke, H, Verma, N, Dollinger, P, Kovacic, F. | | Deposit date: | 2018-06-14 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic insights revealing how lipase binding domain MD1 of Pseudomonas aeruginosa foldase affects lipase activation.
Sci Rep, 10, 2020
|
|
1EX9
 
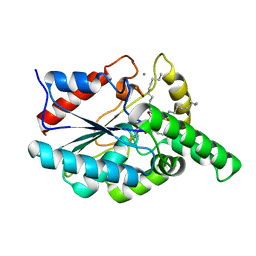 | | CRYSTAL STRUCTURE OF THE PSEUDOMONAS AERUGINOSA LIPASE COMPLEXED WITH RC-(RP,SP)-1,2-DIOCTYLCARBAMOYL-GLYCERO-3-O-OCTYLPHOSPHONATE | | Descriptor: | CALCIUM ION, LACTONIZING LIPASE, OCTYL-PHOSPHINIC ACID 1,2-BIS-OCTYLCARBAMOYLOXY-ETHYL ESTER | | Authors: | Nardini, M, Lang, D.A, Liebeton, K, Jaeger, K.-E, Dijkstra, B.W. | | Deposit date: | 2000-05-02 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of pseudomonas aeruginosa lipase in the open conformation. The prototype for family I.1 of bacterial lipases.
J.Biol.Chem., 275, 2000
|
|
3SW1
 
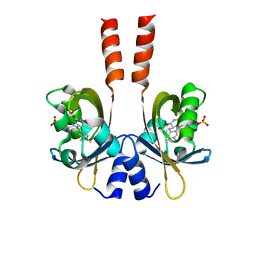 | | Structure of a full-length bacterial LOV protein | | Descriptor: | FLAVIN MONONUCLEOTIDE, Sensory box protein | | Authors: | Granzin, J, Batra-Safferling, R, Jaeger, K.-E, Drepper, T, Krauss, U. | | Deposit date: | 2011-07-13 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural Basis for the Slow Dark Recovery of a Full-Length LOV Protein from Pseudomonas putida.
J.Mol.Biol., 417, 2012
|
|
6Y9K
 
 | |
