1EBE
 
 | | Laue diffraction study on the structure of cytochrome c peroxidase compound I | | Descriptor: | CYTOCHROME C PEROXIDASE, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fulop, V, Phizackerley, R.P, Soltis, S.M, Clifton, I.J, Wakatsuki, S, Erman, J.E, Hajdu, J, Edwards, S.L. | | Deposit date: | 2001-07-25 | | Release date: | 2001-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Laue Diffraction Study on the Structure of Cytochrome C Peroxidase Compound I
Structure, 2, 1994
|
|
1EE3
 
 | | Cadmium-substituted bovine pancreatic carboxypeptidase A (alfa-form) at pH 7.5 and 2 mM chloride in monoclinic crystal form | | Descriptor: | CADMIUM ION, PROTEIN (CARBOXYPEPTIDASE A) | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-01-30 | | Release date: | 2002-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
1ELL
 
 | | CADMIUM-SUBSTITUTED BOVINE PANCREATIC CARBOXYPEPTIDASE A (ALFA-FORM) AT PH 7.5 AND 0.25 M CHLORIDE IN MONOCLINIC CRYSTAL FORM. | | Descriptor: | CADMIUM ION, CARBOXYPEPTIDASE A | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
1ELM
 
 | | CADMIUM-SUBSTITUTED BOVINE PACREATIC CARBOXYPEPTIDASE A (ALFA-FORM) AT PH 5.5 AND 2 MM CHLORIDE IN MONOCLINIC CRYSTAL FORM. | | Descriptor: | CADMIUM ION, CARBOXYPEPTIDASE A | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
1ED3
 
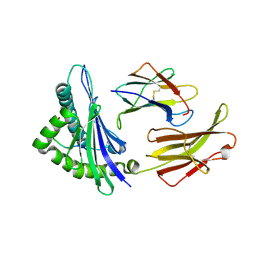 | | CRYSTAL STRUCTURE OF RAT MINOR HISTOCOMPATIBILITY ANTIGEN COMPLEX RT1-AA/MTF-E. | | Descriptor: | BETA-2-MICROGLOBULIN, CLASS I MAJOR HISTOCOMPATIBILITY ANTIGEN RT1-AA, PEPTIDE MTF-E (13N3E) | | Authors: | Speir, J.A, Stevens, J, Joly, E, Butcher, G.W, Wilson, I.A. | | Deposit date: | 2000-01-26 | | Release date: | 2001-02-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Two different, highly exposed, bulged structures for an unusually long peptide bound to rat MHC class I RT1-Aa.
Immunity, 14, 2001
|
|
1EII
 
 | | NMR STRUCTURE OF HOLO CELLULAR RETINOL-BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II, RETINOL | | Authors: | Lu, J, Lin, C.L, Tang, C, Ponder, J.W, Kao, J.L, Cistola, D.P, Li, E. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Binding of retinol induces changes in rat cellular retinol-binding protein II conformation and backbone dynamics.
J.Mol.Biol., 300, 2000
|
|
1EEX
 
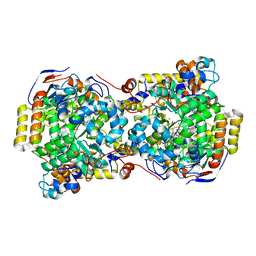 | | CRYSTAL STRUCTURE OF THE DIOL DEHYDRATASE-ADENINYLPENTYLCOBALAMIN COMPLEX FROM KLEBSIELLA OXYTOCA | | Descriptor: | CO-(ADENIN-9-YL-PENTYL)-COBALAMIN, POTASSIUM ION, PROPANEDIOL DEHYDRATASE, ... | | Authors: | Shibata, N, Masuda, J, Toraya, T, Morimoto, Y, Yasuoka, N. | | Deposit date: | 2000-02-04 | | Release date: | 2001-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | How a protein generates a catalytic radical from coenzyme B(12): X-ray structure of a diol-dehydratase-adeninylpentylcobalamin complex.
Structure Fold.Des., 8, 2000
|
|
1ENS
 
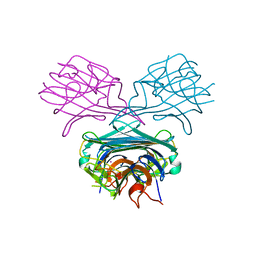 | | CRYSTALS OF DEMETALLIZED CONCANAVALIN A SOAKED WITH COBALT HAVING A COBALT ION BOUND IN THE S1 SITE | | Descriptor: | COBALT (II) ION, CONCANAVALIN A | | Authors: | Bouckaert, J, Loris, R, Poortmans, F, Wyns, L. | | Deposit date: | 1996-03-20 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Sequential structural changes upon zinc and calcium binding to metal-free concanavalin A.
J.Biol.Chem., 271, 1996
|
|
1EGV
 
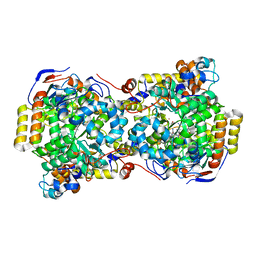 | | CRYSTAL STRUCTURE OF THE DIOL DEHYDRATASE-ADENINYLPENTYLCOBALAMIN COMPLEX FROM KLEBSELLA OXYTOCA UNDER THE ILLUMINATED CONDITION. | | Descriptor: | CO-(ADENIN-9-YL-PENTYL)-COBALAMIN, POTASSIUM ION, PROPANEDIOL DEHYDRATASE, ... | | Authors: | Masuda, J, Shibata, N, Toraya, T, Morimoto, Y, Yasuoka, N. | | Deposit date: | 2000-02-17 | | Release date: | 2001-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | How a protein generates a catalytic radical from coenzyme B(12): X-ray structure of a diol-dehydratase-adeninylpentylcobalamin complex.
Structure Fold.Des., 8, 2000
|
|
1E8K
 
 | | Cyclophilin 3 Complexed With Dipeptide Ala-Pro | | Descriptor: | ALANINE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE 3, PROLINE | | Authors: | Wu, S.Y, Dornan, J, Kontopidis, G, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 2000-09-25 | | Release date: | 2001-09-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The First Direct Determination of a Ligand Binding Constant in Protein Crystals
Angew.Chem.Int.Ed.Engl., 40, 2001
|
|
1EI4
 
 | | B-DNA DODECAMER CGCGAAT(TLC)CGCG WITH INCORPORATED [3.3.0]BICYCLO-ARABINO-THYMINE-5'-PHOSPHATE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(TLC)P*(TLC)P*CP*GP*CP*G)-3'), MAGNESIUM ION, SPERMINE | | Authors: | Minasov, G, Teplova, M, Nielsen, P, Wengel, J, Egli, M. | | Deposit date: | 2000-02-24 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural basis of cleavage by RNase H of hybrids of arabinonucleic acids and RNA.
Biochemistry, 39, 2000
|
|
1EJM
 
 | | CRYSTAL STRUCTURE OF THE BPTI ALA16LEU MUTANT IN COMPLEX WITH BOVINE TRYPSIN | | Descriptor: | BETA-TRYPSIN, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Otlewski, J, Smalas, A, Helland, R, Grzesiak, A, Krowarsch, D. | | Deposit date: | 2000-03-03 | | Release date: | 2001-03-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substitutions at the P(1) position in BPTI strongly affect the association energy with serine proteinases.
J.Mol.Biol., 301, 2000
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4V34
 
 | | The Structure of A-PGS from Pseudomonas aeruginosa (SeMet derivative) | | Descriptor: | ALANYL-TRNA-DEPENDENT L-ALANYL- PHOPHATIDYLGLYCEROL SYNTHASE, CHLORIDE ION, SULFATE ION | | Authors: | Krausze, J, Hebecker, S, Hasenkampf, T, Heinz, D.W, Moser, J. | | Deposit date: | 2014-10-16 | | Release date: | 2015-08-19 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Two Bacterial Resistance Factors Mediating tRNA-Dependent Aminoacylation of Phosphatidylglycerol with Lysine or Alanine.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4V35
 
 | | The Structure of A-PGS from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, ALANYL-TRNA-DEPENDENT L-ALANYL- PHOPHATIDYLGLYCEROL SYNTHASE, CALCIUM ION, ... | | Authors: | Krausze, J, Hebecker, S, Hasenkampf, T, Heinz, D.W, Moser, J. | | Deposit date: | 2014-10-16 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Two Bacterial Resistance Factors Mediating tRNA-Dependent Aminoacylation of Phosphatidylglycerol with Lysine or Alanine.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X5Y
 
 | | Menin in complex with MI-503 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-methyl-1-(1H-pyrazol-4-ylmethyl)-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, ... | | Authors: | Pollock, J, Borkin, D, Cierpicki, T, Grembecka, J. | | Deposit date: | 2014-12-06 | | Release date: | 2015-04-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Pharmacologic Inhibition of the Menin-MLL Interaction Blocks Progression of MLL Leukemia In Vivo.
Cancer Cell, 27, 2015
|
|
4X5Z
 
 | | menin in complex with MI-136 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Borkin, D, Cierpicki, T, Grembecka, J. | | Deposit date: | 2014-12-06 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pharmacologic Inhibition of the Menin-MLL Interaction Blocks Progression of MLL Leukemia In Vivo.
Cancer Cell, 27, 2015
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4WOT
 
 | | ROCK2 IN COMPLEX WITH 1426382-07-1 | | Descriptor: | Rho-associated protein kinase 2, methyl 3-[({2'-(aminomethyl)-5'-[(3-fluoropyridin-4-yl)carbamoyl]biphenyl-3-yl}carbonyl)amino]-4-fluorobenzoate | | Authors: | Augustin, M, Krapp, S, Boland, S, Defert, O, Bourin, A, Alen, J, Leysen, D. | | Deposit date: | 2014-10-16 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Design, synthesis, and biological evaluation of novel, highly active soft ROCK inhibitors.
J. Med. Chem., 58, 2015
|
|
4X1T
 
 | | The crystal structure of Arabidopsis thaliana galactolipid synthase MGD1 in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, Monogalactosyldiacylglycerol synthase 1, chloroplastic, ... | | Authors: | Rocha, J, Breton, C. | | Deposit date: | 2014-11-25 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights and membrane binding properties of MGD1, the major galactolipid synthase in plants.
Plant J., 85, 2016
|
|
4WYI
 
 | |
4YWC
 
 | | Crystal structure of Myc3(5-242) fragment in complex with Jaz9(218-239) peptide | | Descriptor: | Protein TIFY 7, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2015-03-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4YNE
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 6-[(2R)-2-(3-fluorophenyl)pyrrolidin-1-yl]-3-(pyridin-2-yl)imidazo[1,2-b]pyridazine, GLYCEROL, High affinity nerve growth factor receptor, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0229 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4YMJ
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-[6-(benzylamino)imidazo[1,2-b]pyridazin-3-yl]benzonitrile, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-06 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
