5ZRU
 
 | | Crystal structure of Agl-KA catalytic domain | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Yano, S, Makabe, K. | | Deposit date: | 2018-04-25 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Crystal structure of the catalytic unit of GH 87-type alpha-1,3-glucanase Agl-KA from Bacillus circulans.
Sci Rep, 9, 2019
|
|
6Z2J
 
 | | The structure of the dimeric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
6Z2K
 
 | | The structure of the tetrameric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
2ZYZ
 
 | | Pyrobaculum aerophilum splicing endonuclease | | Descriptor: | Putative uncharacterized protein PAE0789, tRNA-splicing endonuclease | | Authors: | Yoshinari, S, Inaoka, D.K, Watanabe, Y, Shiba, T, Kurisu, G, Harada, S. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional importance of crenarchaea-specific extra-loop revealed by an X-ray structure of a heterotetrameric crenarchaeal splicing endonuclease
Nucleic Acids Res., 37, 2009
|
|
2ZYC
 
 | | Crystal structure of peptidoglycan hydrolase from Sphingomonas sp. A1 | | Descriptor: | PHOSPHATE ION, Peptidoglycan hydrolase FlgJ | | Authors: | Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-01-19 | | Release date: | 2009-02-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the glycosidase family 73 peptidoglycan hydrolase FlgJ
Biochem.Biophys.Res.Commun., 381, 2009
|
|
2ZZR
 
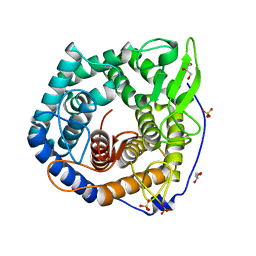 | |
6ZLY
 
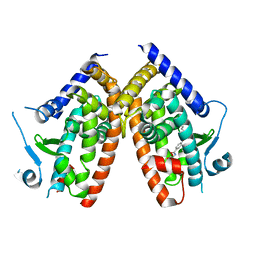 | |
2E8Z
 
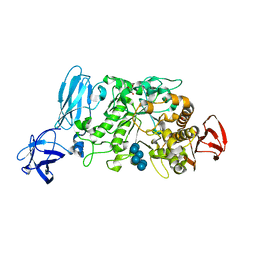 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 complexed with alpha-cyclodextrin | | Descriptor: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | Authors: | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 in complex with maltose and alpha-cyclodextrin
To be Published
|
|
2E9B
 
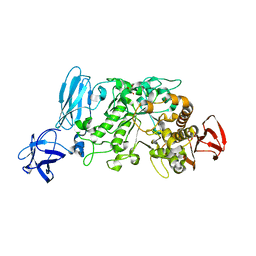 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 complexed with maltose | | Descriptor: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | Authors: | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 in complex with maltose and alpha-cyclodextrin
To be Published
|
|
2E8Y
 
 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 | | Descriptor: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | Authors: | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of pullulanase type I from Bacillus subtilis str. 168
in complex with maltose and alpha-cyclodextrin
To be Published
|
|
