5JKC
 
 | | Crystal structure of human IZUMO1-JUNO complex (crystal form 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Izumo sperm-egg fusion protein 1, ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2016-04-26 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of IZUMO1-JUNO reveals sperm-oocyte recognition during mammalian fertilization
Nature, 534, 2016
|
|
5JKD
 
 | | Crystal structure of human IZUMO1-JUNO complex (crystal form 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Izumo sperm-egg fusion protein 1, ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2016-04-26 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of IZUMO1-JUNO reveals sperm-oocyte recognition during mammalian fertilization
Nature, 534, 2016
|
|
5JKE
 
 | | Crystal structure of human IZUMO1-JUNO complex (crystal form 3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Izumo sperm-egg fusion protein 1, ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2016-04-26 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of IZUMO1-JUNO reveals sperm-oocyte recognition during mammalian fertilization
Nature, 534, 2016
|
|
5JKB
 
 | |
5JKA
 
 | | Crystal structure of human JUNO (crystal form 1) | | Descriptor: | CHLORIDE ION, Sperm-egg fusion protein Juno, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2016-04-26 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of IZUMO1-JUNO reveals sperm-oocyte recognition during mammalian fertilization
Nature, 534, 2016
|
|
2KSZ
 
 | |
1G01
 
 | | ALKALINE CELLULASE K CATALYTIC DOMAIN | | Descriptor: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE | | Authors: | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | Deposit date: | 2000-10-05 | | Release date: | 2001-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
1G0C
 
 | | ALKALINE CELLULASE K CATALYTIC DOMAIN-CELLOBIOSE COMPLEX | | Descriptor: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE, ... | | Authors: | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | Deposit date: | 2000-10-05 | | Release date: | 2001-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
2LX9
 
 | |
2M7E
 
 | |
2N98
 
 | | Solution structure of acyl carrier protein LipD from Actinoplanes friuliensis | | Descriptor: | Acyl carrier protein | | Authors: | Paul, S, Ishida, H, Liu, Z, Nguyen, L.T, Vogel, H.J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-11-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic characterization of a freestanding acyl carrier protein involved in the biosynthesis of cyclic lipopeptide antibiotics.
Protein Sci., 26, 2017
|
|
2LGF
 
 | |
2L7L
 
 | |
2M73
 
 | |
3AXM
 
 | | Structure of rice Rubisco in complex with 6PG | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Matsumura, H, Mizohata, E, Ishida, H, Kogami, A, Ueno, T, Makino, A, Inoue, T, Yokota, A, Mae, T, Kai, Y. | | Deposit date: | 2011-04-11 | | Release date: | 2012-04-11 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
3AXK
 
 | | Structure of rice Rubisco in complex with NADP(H) | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Matsumura, H, Mizohata, E, Ishida, H, Kogami, A, Ueno, T, Makino, A, Inoue, T, Yokota, A, Mae, T, Kai, Y. | | Deposit date: | 2011-04-11 | | Release date: | 2012-04-11 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
2ZFD
 
 | | The crystal structure of plant specific calcium binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14 | | Descriptor: | ACETIC ACID, CALCIUM ION, Calcineurin B-like protein 2, ... | | Authors: | Akaboshi, M, Hashimoto, H, Ishida, H, Koizumi, N, Sato, M, Shimizu, T. | | Deposit date: | 2007-12-29 | | Release date: | 2008-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of plant-specific calcium-binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14
J.Mol.Biol., 377, 2008
|
|
5Y3J
 
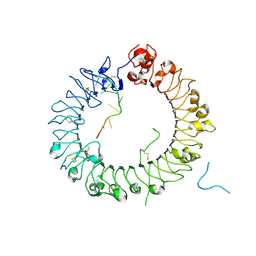 | | Crystal structure of horse TLR9 in complex with two DNAs (CpG DNA and TCGCAC DNA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*AP*GP*GP*CP*GP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2017-07-29 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Toll-like Receptor 9 Contains Two DNA Binding Sites that Function Cooperatively to Promote Receptor Dimerization and Activation
Immunity, 48, 2018
|
|
3WLU
 
 | | Crystal Structure of human galectin-9 NCRD with Selenolactose | | Descriptor: | 2-(trimethylsilyl)ethyl 4-O-beta-D-galactopyranosyl-6-Se-methyl-6-seleno-beta-D-glucopyranoside, Galectin-9 | | Authors: | Makyio, H, Suzuki, T, Ando, H, Yamada, Y, Ishida, H, Kiso, M, Wakatsuki, S, Kato, R. | | Deposit date: | 2013-11-14 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expanded potential of seleno-carbohydrates as a molecular tool for X-ray structural determination of a carbohydrate-protein complex with single/multi-wavelength anomalous dispersion phasing
Bioorg.Med.Chem., 22, 2014
|
|
7EFQ
 
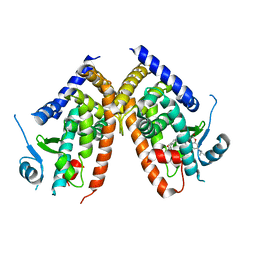 | | Crystal structure of hPPARgamma ligand binding domain complexed with rosiglitazone-based fluorescence probe | | Descriptor: | (5S)-5-[[4-[2-[[7-(diethylamino)-2-oxidanylidene-chromen-4-yl]-methyl-amino]ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Yoshikawa, C, Ishida, H, Ohashi, N, Itoh, T. | | Deposit date: | 2021-03-23 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis of a Coumarin-Based PPAR gamma Fluorescence Probe for Competitive Binding Assay.
Int J Mol Sci, 22, 2021
|
|
3WVM
 
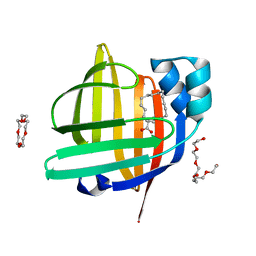 | | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with stearic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Matsumura, H, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-25 | | Release date: | 2015-01-28 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type fatty-acid-binding protein.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3WBG
 
 | | Structure of the human heart fatty acid-binding protein in complex with 1-anilinonaphtalene-8-sulphonic acid | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, Fatty acid-binding protein, heart | | Authors: | Hirose, M, Sugiyama, S, Ishida, H, Niiyama, M, Matsuoka, D, Hara, T, Sato, F, Mizohata, E, Murakami, S, Inoue, T, Matsuoka, S, Murata, M. | | Deposit date: | 2013-05-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the human-heart fatty-acid-binding protein 3 in complex with the fluorescent probe 1-anilinonaphthalene-8-sulphonic acid
J.SYNCHROTRON RADIAT., 20, 2013
|
|
3VJL
 
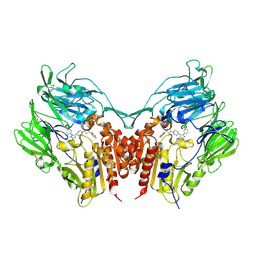 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with a prolylthiazolidine inhibitor #2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Discovery and preclinical profile of teneligliptin (3-[(2S,4S)-4-[4-(3-methyl-1-phenyl-1H-pyrazol-5-yl)piperazin-1-yl]pyrrolidin-2-ylcarbonyl]thiazolidine): A highly potent, selective, long-lasting and orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem., 20, 2012
|
|
3VJK
 
 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with MP-513 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery and preclinical profile of teneligliptin (3-[(2S,4S)-4-[4-(3-methyl-1-phenyl-1H-pyrazol-5-yl)piperazin-1-yl]pyrrolidin-2-ylcarbonyl]thiazolidine): A highly potent, selective, long-lasting and orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem., 20, 2012
|
|
8HB0
 
 | | Structure of human SGLT2-MAP17 complex with TA1887 | | Descriptor: | (2R,3R,4S,5S,6R)-2-[3-[(4-cyclopropylphenyl)methyl]-4-fluoranyl-indol-1-yl]-6-(hydroxymethyl)oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, PDZK1-interacting protein 1, ... | | Authors: | Hiraizumi, M, Kishida, H, Miyaguchi, I, Nureki, O. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Transport and inhibition mechanism of the human SGLT2-MAP17 glucose transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
