4IEA
 
 | |
6DSS
 
 | | Re-refinement of P. falciparum orotidine 5'-monophosphate decarboxylase | | Descriptor: | Orotidine 5'-monophosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Brandt, G.S, Novak, W.R.P. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Re-refinement of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase provides a clearer picture of an important malarial drug target.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6DSQ
 
 | |
6DSR
 
 | | Re-refinement of P. falciparum orotidine 5'-monophosphate decarboxylase | | Descriptor: | Orotidine 5'-monophosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Brandt, G.S, Novak, W.R.P. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Re-refinement of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase provides a clearer picture of an important malarial drug target.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3R8R
 
 | |
1JQO
 
 | |
1JQN
 
 | |
8GZW
 
 | | Klebsiella pneumoniae FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZV
 
 | | Klebsiella pneumoniae FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZX
 
 | | Escherichia coli FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZY
 
 | | Escherichia coli FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
1H8A
 
 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX3 | | Descriptor: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-31 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1H89
 
 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX2 | | Descriptor: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-30 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
4YMY
 
 | | Crystal structure of mutant nitrobindin M75A/H76L/Q96C/M148L/H158A (NB11) from Arabidopsis thaliana | | Descriptor: | GLYCEROL, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Mizohata, E, Himiyama, T, Tachikawa, K, Oohora, K, Onoda, A, Hayashi, T. | | Deposit date: | 2015-03-08 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A Highly Active Biohybrid Catalyst for Olefin Metathesis in Water: Impact of a Hydrophobic Cavity in a beta-Barrel Protein
Acs Catalysis, 5, 2015
|
|
1H88
 
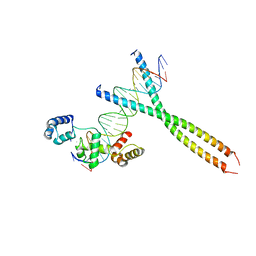 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX1 | | Descriptor: | AMMONIUM ION, CCAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-29 | | Release date: | 2002-01-28 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1IO4
 
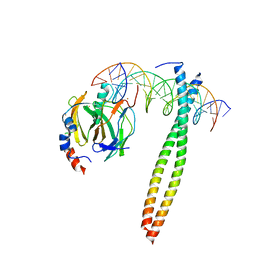 | |
5B3F
 
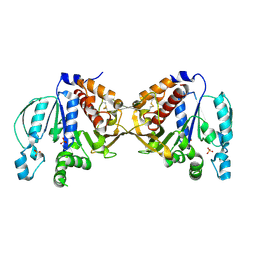 | |
7E5O
 
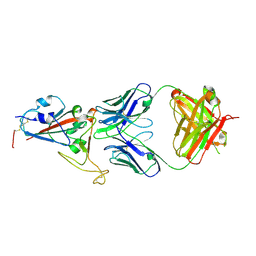 | | Crystal structure of SARS-CoV-2 RBD in complex with antibody NT-193 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-193 Heavy chain, NT-193 Light chain, ... | | Authors: | Kita, S, Onodera, T, Adachi, Y, Moriayma, S, Nomura, T, Tadokoro, T, Anraku, Y, Yumoto, K, Tian, C, Fukuhara, H, Suzuki, T, Tonouchi, K, Sasaki, J, Sun, L, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 antibody broadly neutralizes SARS-related coronaviruses and variants by coordinated recognition of a virus-vulnerable site.
Immunity, 54, 2021
|
|
3KKQ
 
 | | Crystal structure of M-Ras P40D in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein M-Ras | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3KKM
 
 | | Crystal structure of H-Ras T35S in complex with GppNHp | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3KKP
 
 | | Crystal structure of M-Ras P40D in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein M-Ras | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3KKN
 
 | | Crystal structure of H-Ras T35S in complex with GppNHp | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3KKO
 
 | | Crystal structure of M-Ras P40D/D41E/L51R in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein M-Ras, ... | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
7EA6
 
 | | Crystal structure of TCR-017 ectodomain | | Descriptor: | T cell receptor 017 alpha chain, T cell receptor 017 beta chain | | Authors: | Nagae, M, Yamasaki, S. | | Deposit date: | 2021-03-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18000245 Å) | | Cite: | Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients.
J.Exp.Med., 218, 2021
|
|
5AX9
 
 | | Crystal structure of the kinase domain of human TRAF2 and NCK-interacting protein kinase in complex with compund 9 | | Descriptor: | 4-methoxy-3-[2-[(3-methoxy-4-morpholin-4-yl-phenyl)amino]pyridin-4-yl]benzenecarbonitrile, SULFATE ION, TRAF2 and NCK-interacting protein kinase | | Authors: | Ohbayashi, N, Kukimoto-Niino, M, Yamada, T, Shirouzu, M. | | Deposit date: | 2015-07-21 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | TNIK inhibition abrogates colorectal cancer stemness
Nat Commun, 7, 2016
|
|
