2Q33
 
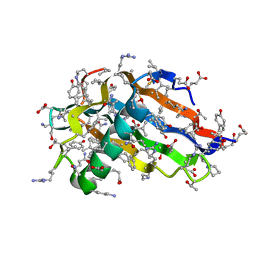 | | Crystal structure of all-D monellin at 1.8 A resolution | | Descriptor: | D-MONELLIN CHAIN A, D-MONELLIN CHAIN B | | Authors: | Hung, L.-W, Kohmura, M, Ariyoshi, Y, Kim, S.-H. | | Deposit date: | 2007-05-29 | | Release date: | 2007-11-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an Enantiomeric Protein, D-Monellin at 1.8 A Resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
8DJ8
 
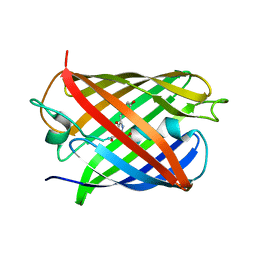 | | Crystal Structure of Calgreen 1 protein | | Descriptor: | Calgreen One | | Authors: | Hung, L.-W, Nguyen, H.B, Waldo, G. | | Deposit date: | 2022-06-30 | | Release date: | 2023-07-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of Engineered Variants of GFP-type Green Fluorescent protein from Corynactis californica (tentative)
To Be Published
|
|
4LMP
 
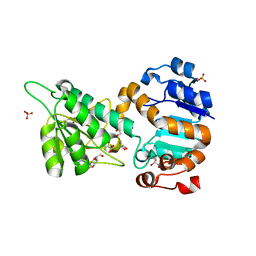 | | Mycobacterium tuberculosis L-alanine dehydrogenase x-ray structure in complex with N6-methyl adenosine | | Descriptor: | Alanine dehydrogenase, GLYCEROL, N-methyladenosine, ... | | Authors: | Kim, H.-B, Hung, L.-W, Goulding, C.W, Terwilliger, T.C, Kim, C.-Y, Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-07-10 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Drug target analysis by dye-ligand affinity chromatography
To be Published
|
|
1SBZ
 
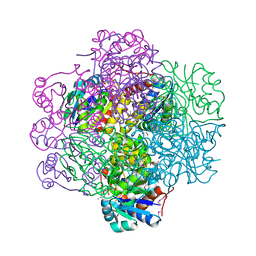 | | Crystal Structure of dodecameric FMN-dependent Ubix-like Decarboxylase from Escherichia coli O157:H7 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable aromatic acid decarboxylase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Tocilj, A, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-02-11 | | Release date: | 2004-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a dodecameric FMN-dependent UbiX-like decarboxylase (Pad1) from Escherichia coli O157: H7.
Protein Sci., 13, 2004
|
|
3LP6
 
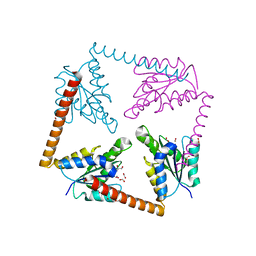 | | Crystal Structure of Rv3275c-E60A from Mycobacterium tuberculosis at 1.7A resolution | | Descriptor: | FORMIC ACID, GLYCEROL, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Kim, H, Yu, M, Hung, L.-W, Terwilliger, T.C, Kim, C.-Y, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal Structure of Rv3275c-E60A from Mycobacterium tuberculosis at 1.7A
Resolution
To be Published
|
|
2A2J
 
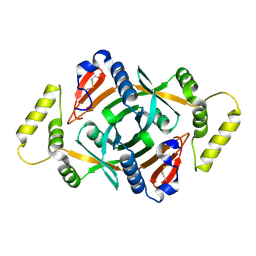 | | Crystal structure of a putative pyridoxine 5'-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis | | Descriptor: | Pyridoxamine 5'-phosphate oxidase | | Authors: | Pedelacq, J.-D, Rho, B.-S, Kim, C.-Y, Waldo, G.S, Lekin, T.P, Segelke, B.W, Rupp, B, Hung, L.-W, Kim, S.-I, Terwilliger, T.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-06-22 | | Release date: | 2005-08-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative pyridoxine 5'-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis.
Proteins, 62, 2005
|
|
3B4W
 
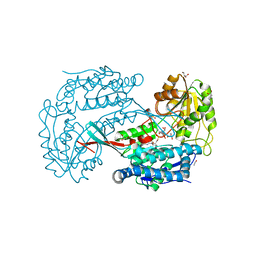 | | Crystal structure of Mycobacterium tuberculosis aldehyde dehydrogenase complexed with NAD+ | | Descriptor: | Aldehyde dehydrogenase, ETHANOL, GLYCEROL, ... | | Authors: | Moon, J.H, Lyon, A.E, Yu, M, Hung, L.-W, Terwilliger, T, Kim, C.-Y, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-10-24 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of aldehyde dehydrogenase from Mycobacterium tuberculosis complexed with NAD+.
To Be Published
|
|
4HC6
 
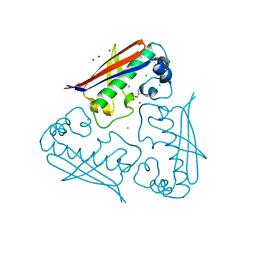 | | Mycobacterium tuberculosis Rv2523cE77A x-ray structure solved with 1.8 angstrom resolution | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, H.-B, Han, G.-W, Hung, L.-W, Terwilliger, C.T, Kim, C.-Y, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2012-09-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mycobacterium tuberculosis Rv2523cE77A x-ray structure solved with 1.8 angstrom resolution
To be Published
|
|
6O7F
 
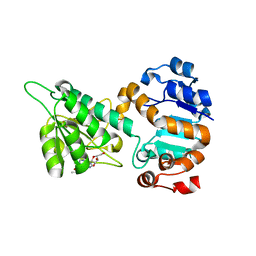 | |
1N3B
 
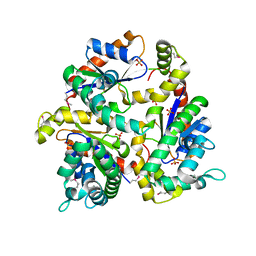 | | Crystal Structure of Dephosphocoenzyme A kinase from Escherichia coli | | Descriptor: | Dephospho-CoA kinase, SULFATE ION | | Authors: | O'Toole, N, Barbosa, J.A.R.G, Li, Y, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Trimeric Form of Dephosphocoenzyme A Kinase from Escherichia coli
Protein Sci., 12, 2003
|
|
1MJH
 
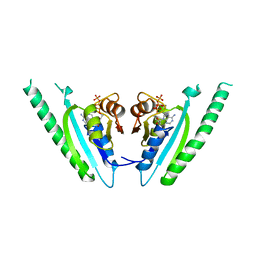 | | Structure-based assignment of the biochemical function of hypothetical protein MJ0577: A test case of structural genomics | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PROTEIN (ATP-BINDING DOMAIN OF PROTEIN MJ0577) | | Authors: | Zarembinski, T.I, Hung, L.-W, Mueller-Dieckmann, H.J, Kim, K.-K, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1998-11-04 | | Release date: | 1998-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based assignment of the biochemical function of a hypothetical protein: a test case of structural genomics.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2NYX
 
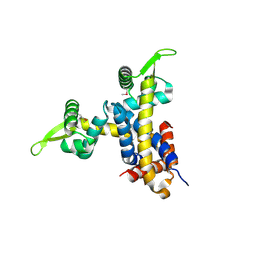 | | Crystal structure of RV1404 from Mycobacterium tuberculosis | | Descriptor: | Probable transcriptional regulatory protein, Rv1404 | | Authors: | Yu, M, Bursey, E.H, Radhakannan, R, Kim, C.-Y, Kaviratne, T, Woodruff, T, Segelke, B.W, Lekin, T, Toppani, D, Terwilliger, T.C, Hung, L.-W, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of RV1404 from Mycobacterium tuberculosis
To be Published
|
|
4JJT
 
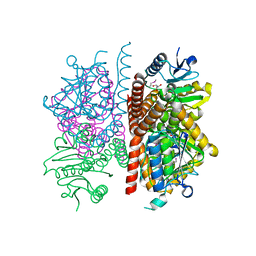 | | The crystal structure of enoyl-CoA hydratase from Mycobacterium tuberculosis H37Rv | | Descriptor: | ACETATE ION, Enoyl-CoA hydratase, GLYCEROL | | Authors: | Tan, K, Holowicki, J, Endres, M, Kim, C.-Y, Kim, H, Hung, L.-W, Terwilliger, T.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-03-08 | | Release date: | 2013-03-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | The crystal structure of enoyl-CoA hydratase from Mycobacterium tuberculosis H37Rv
To be Published
|
|
4JBM
 
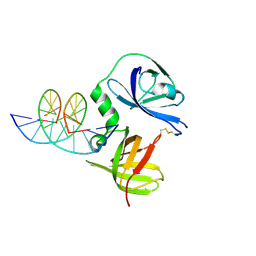 | | Structure of murine DNA binding protein bound with ds DNA | | Descriptor: | DNA (5'-D(*GP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), Interferon-inducible protein AIM2 | | Authors: | Ru, H, Ni, X, Crowley, C, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
4JBK
 
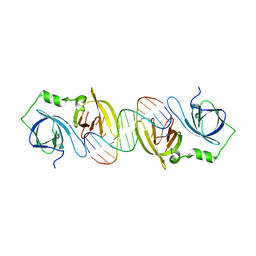 | | Molecular basis for abrogation of activation of pro-inflammatory cytokines | | Descriptor: | DNA (5'-D(P*GP*GP*AP*AP*TP*TP*AP*TP*AP*AP*TP*TP*CP*C)-3'), Interferon-activable protein 202 | | Authors: | Ru, H, Ni, X, Crowley, C, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
2FSX
 
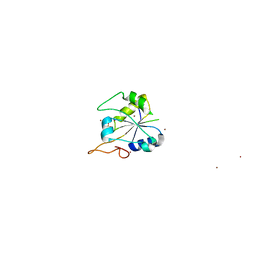 | | Crystal structure of Rv0390 from M. tuberculosis | | Descriptor: | BROMIDE ION, COG0607: Rhodanese-related sulfurtransferase, SULFATE ION | | Authors: | Bursey, E.H, Radhakannan, T, Yu, M, Segelke, B.W, Lekin, T, Toppani, D, Chang, Y.-B, Kaviratne, T, Woodruff, T, Terwilliger, T.C, Hung, L.-W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-01-23 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Rv0390 from Mycobacterium tuberculosis
To be Published
|
|
2F0A
 
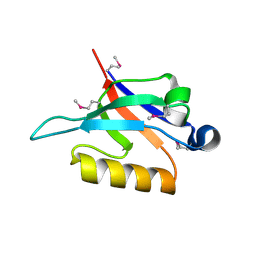 | | Crystal Structure of Monomeric Uncomplexed form of Xenopus dishevelled PDZ domain | | Descriptor: | COBALT (II) ION, SULFATE ION, Segment polarity protein dishevelled homolog DVL-2 | | Authors: | Friedland, N, Hung, L.-W, Cheyette, B, Moon, R.T, Earnest, T.N. | | Deposit date: | 2005-11-12 | | Release date: | 2005-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational flexibility in the PDZ domain of Dishevelled induced by target binding
TO BE PUBLISHED
|
|
4JBJ
 
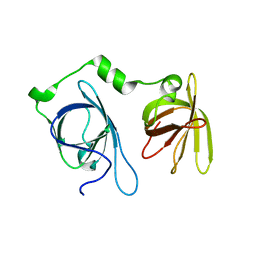 | | Structural mimicry for functional antagonism | | Descriptor: | Interferon-activable protein 202 | | Authors: | Ru, H, Ni, X, Ma, F, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
2G2D
 
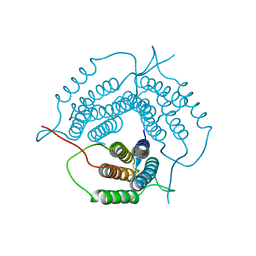 | | Crystal structure of a putative pduO-type ATP:cobalamin adenosyltransferase from Mycobacterium tuberculosis | | Descriptor: | ATP:cobalamin adenosyltransferase | | Authors: | Moon, J.H, Kaviratne, A, Yu, M, Bursey, E.H, Hung, L.-W, Lekin, T.P, Segelke, B.W, Terwilliger, T.C, Kim, C.-Y, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-02-15 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative pduO-type ATP:cobalamin adenosyltransferase from Mycobacterium tuberculosis.
To be Published
|
|
1DUQ
 
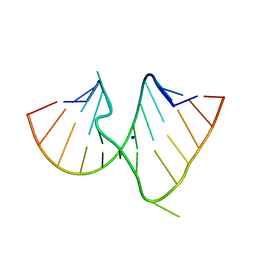 | |
1B0U
 
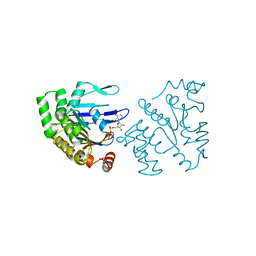 | | ATP-BINDING SUBUNIT OF THE HISTIDINE PERMEASE FROM SALMONELLA TYPHIMURIUM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, HISTIDINE PERMEASE | | Authors: | Hung, L.-W, Wang, I.X, Nikaido, K, Liu, P.-Q, Ames, G.F.-L, Kim, S.-H. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the ATP-binding subunit of an ABC transporter.
Nature, 396, 1998
|
|
8U23
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 8 | | Descriptor: | Green Fluorescent Protein Variant #8, ccGFP 8 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U20
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 5 | | Descriptor: | Green Fluorescent Protein Variant #5, ccGFP 5 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U22
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 7 | | Descriptor: | Green Fluorescent Protein Variant #7, ccGFP 7 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U21
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP E6 | | Descriptor: | Green Fluorescent Protein Variant E6, ccGFP E6 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
