1U32
 
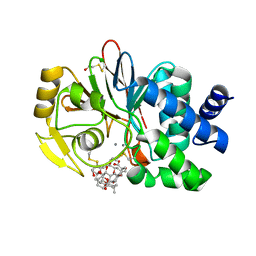 | | Crystal structure of a Protein Phosphatase-1: Calcineurin Hybrid Bound to Okadaic Acid | | Descriptor: | BETA-MERCAPTOETHANOL, MANGANESE (II) ION, OKADAIC ACID, ... | | Authors: | Maynes, J.T, Perreault, K.R, Cherney, M.M, Luu, H.A, James, M.N.G, Holmes, C.F.B. | | Deposit date: | 2004-07-20 | | Release date: | 2004-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Mutagenesis of a Protein Phosphatase-1:Calcineurin Hybrid Elucidate the Role of the {beta}12-{beta}13 Loop in Inhibitor Binding
J.Biol.Chem., 279, 2004
|
|
6DCX
 
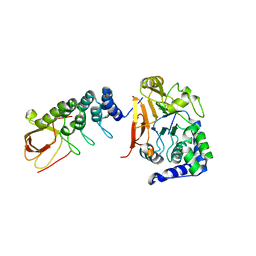 | | iASPP-PP-1c structure and targeting of p53 | | Descriptor: | RelA-associated inhibitor, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Glover, J.N.M, Zhou, Y, Edwards, R.A. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure, 27, 2019
|
|
1EVC
 
 | |
1EVD
 
 | |
