3WFU
 
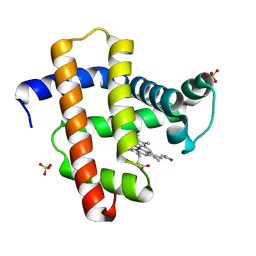 | | Crystal structure of horse heart myoglobin reconstituted with cobalt(I) tetradehydrocorrin | | Descriptor: | (1R,19R) cobalt tetradehydrocorrin, (1S,19S) cobalt tetradehydrocorrin, Myoglobin, ... | | Authors: | Mizohata, E, Morita, Y, Oohora, K, Hirata, K, Ohbayashi, J, Inoue, T, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2013-07-23 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Co(II)/Co(I) reduction-induced axial histidine-flipping in myoglobin reconstituted with a cobalt tetradehydrocorrin as a methionine synthase model.
Chem.Commun.(Camb.), 50, 2014
|
|
3WFT
 
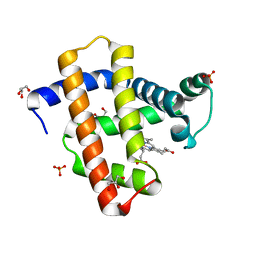 | | Crystal structure of horse heart myoglobin reconstituted with cobalt(II) tetradehydrocorrin | | Descriptor: | (1R,19R) cobalt tetradehydrocorrin, (1S,19S) cobalt tetradehydrocorrin, GLYCEROL, ... | | Authors: | Mizohata, E, Morita, Y, Oohora, K, Hirata, K, Ohbayashi, J, Inoue, T, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2013-07-23 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Co(II)/Co(I) reduction-induced axial histidine-flipping in myoglobin reconstituted with a cobalt tetradehydrocorrin as a methionine synthase model.
Chem.Commun.(Camb.), 50, 2014
|
|
2D6C
 
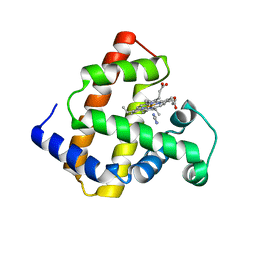 | | Crystal structure of myoglobin reconstituted with iron porphycene | | Descriptor: | IMIDAZOLE, Myoglobin, PORPHYCENE CONTAINING FE | | Authors: | Hayashi, T, Murata, D, Makino, M, Sugimoto, H, Matsuo, T, Sato, H, Shiro, Y, Hisaeda, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-11 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure and peroxidase activity of myoglobin reconstituted with iron porphycene
Inorg.Chem., 45, 2006
|
|
2EKT
 
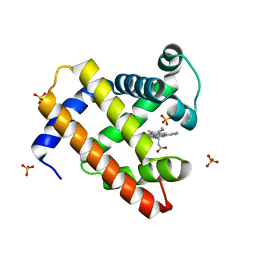 | | Crystal structure of myoglobin reconstituted with 6-methyl-6-depropionatehemin | | Descriptor: | 6-METHY-6-DEPROPIONATEHEMIN, Myoglobin, SULFATE ION | | Authors: | Harada, K, Makino, M, Sugimoto, H, Hirota, S, Matsuo, T, Shiro, Y, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2007-03-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and ligand binding properties of myoglobins reconstituted with monodepropionated heme: functional role of each heme propionate side chain
Biochemistry, 46, 2007
|
|
2EKU
 
 | | Crystal structure of myoglobin reconstituted with 7-methyl-7-depropionatehemin | | Descriptor: | 7-METHYL-7-DEPROPIONATEHEMIN, Myoglobin, SULFATE ION | | Authors: | Harada, K, Makino, M, Sugimoto, H, Hirota, S, Matsuo, T, Shiro, Y, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2007-03-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and ligand binding properties of myoglobins reconstituted with monodepropionated heme: functional role of each heme propionate side chain
Biochemistry, 46, 2007
|
|
6JLG
 
 | | Crystal Structure of HasAp with Co-9,10,19,20-Tetraphenylporphycene | | Descriptor: | GLYCEROL, Heme acquisition protein HasA, PHOSPHATE ION, ... | | Authors: | Sakakibara, E, Shisaka, Y, Onoda, H, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-03-05 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly malleable haem-binding site of the haemoprotein HasA permits stable accommodation of bulky tetraphenylporphycenes.
Rsc Adv, 9, 2019
|
|
7CD0
 
 | | Crystal structure of the 2-iodoporphobilinogen-bound ES2 intermediate form of human hydroxymethylbilane synthase | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, 3-[5-(aminomethyl)-4-(carboxymethyl)-2-iodo-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
7CCZ
 
 | | Crystal structure of the ES2 intermediate form of human hydroxymethylbilane synthase | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
7CCX
 
 | | Crystal structure of the holo form of human hydroxymethylbilane synthase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
7CCY
 
 | | Crystal structure of the 2-iodoporphobilinogen-bound holo form of human hydroxymethylbilane synthase | | Descriptor: | 3-[5-(aminomethyl)-4-(carboxymethyl)-2-iodo-1H-pyrrol-3-yl]propanoic acid, 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
