2RQ6
 
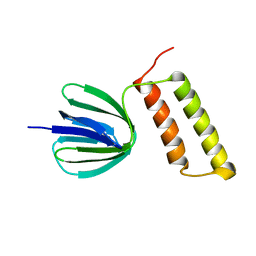 | | Solution structure of the epsilon subunit of the F1-atpase from thermosynechococcus elongatus BP-1 | | Descriptor: | ATP synthase epsilon chain | | Authors: | Yagi, H, Konno, H, Murakami-Fuse, T, Oroguchi, H, Akutsu, T, Ikeguchi, M, Hisabori, T. | | Deposit date: | 2009-03-03 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of the intrinsic inhibitor subunit epsilon of F1-ATPase from photosynthetic organisms.
Biochem.J., 425, 2010
|
|
2RQ7
 
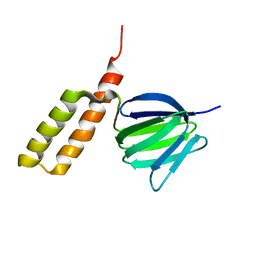 | | Solution structure of the epsilon subunit chimera combining the N-terminal beta-sandwich domain from T. Elongatus bp-1 f1 and the C-terminal alpha-helical domain from spinach chloroplast F1 | | Descriptor: | ATP synthase epsilon chain,ATP synthase epsilon chain, chloroplastic | | Authors: | Yagi, H, Konno, H, Murakami-Fuse, T, Oroguchi, H, Akutsu, T, Ikeguchi, M, Hisabori, T. | | Deposit date: | 2009-03-03 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of the intrinsic inhibitor subunit epsilon of F1-ATPase from photosynthetic organisms.
Biochem.J., 425, 2010
|
|
6AA2
 
 | | X-ray structure of ReQy1 (oxidized form) | | Descriptor: | Green fluorescent protein | | Authors: | Sugiura, K, Yasuda, A, Tabushi, N, Tanaka, H, Kurisu, G, Hisabori, T. | | Deposit date: | 2018-07-17 | | Release date: | 2019-05-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multicolor redox sensor proteins can visualize redox changes in various compartments of the living cell.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
5ZWL
 
 | |
3AFV
 
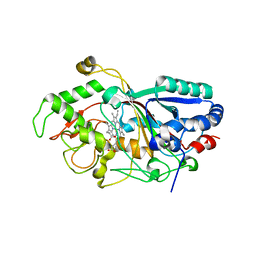 | | Dye-decolorizing peroxidase (DyP) at 1.4 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DyP, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugano, Y, Yoshida, T, Tsuge, H. | | Deposit date: | 2010-03-11 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The catalytic mechanism of dye-decolorizing peroxidase DyP may require the swinging movement of an aspartic acid residue
Febs J., 278, 2011
|
|
5C2I
 
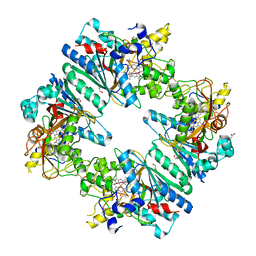 | | Crystal structure of Anabaena sp. DyP-type peroxidese (AnaPX) | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Alr1585 protein, ... | | Authors: | Yoshida, T, Amano, Y, Tsuge, H, Sugano, Y. | | Deposit date: | 2015-06-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Anabaena sp. DyP-type peroxidase is a tetramer consisting of two asymmetric dimers.
Proteins, 84, 2016
|
|
2D3Q
 
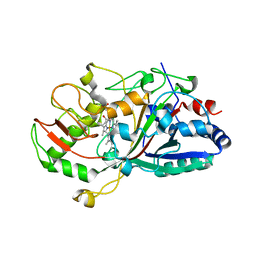 | |
3MM2
 
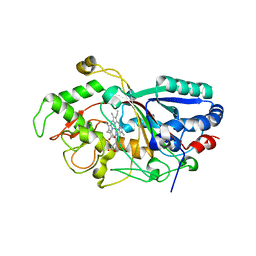 | | Dye-decolorizing peroxidase (DyP) in complex with cyanide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYANIDE ION, DyP, ... | | Authors: | Sugano, Y, Yoshida, T, Tsuge, H. | | Deposit date: | 2010-04-19 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The catalytic mechanism of dye-decolorizing peroxidase DyP may require the swinging movement of an aspartic acid residue
Febs J., 278, 2011
|
|
3MM1
 
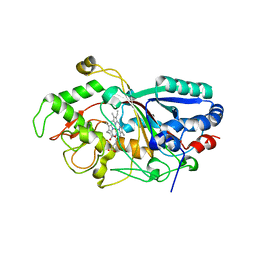 | | Dye-decolorizing peroxidase (DyP) D171N | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DyP, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugano, Y, Yoshida, T, Tsuge, H. | | Deposit date: | 2010-04-19 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The catalytic mechanism of dye-decolorizing peroxidase DyP may require the swinging movement of an aspartic acid residue
Febs J., 278, 2011
|
|
3MM3
 
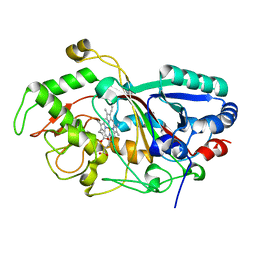 | |
6AA6
 
 | |
7C2B
 
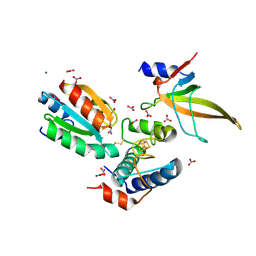 | | Crystal structure of ferredoxin: thioredoxin reductase and thioredoxin f2 complex | | Descriptor: | Ferredoxin-thioredoxin reductase catalytic chain, chloroplastic, Ferredoxin-thioredoxin reductase variable chain, ... | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7949 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
7BZK
 
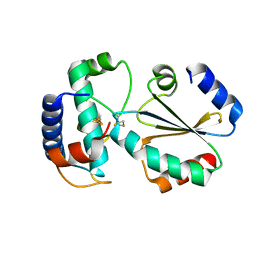 | | Crystal structure of ferredoxin: thioredoxin reductase and thioredoxin y1 complex | | Descriptor: | Ferredoxin-thioredoxin reductase catalytic chain, chloroplastic, IRON/SULFUR CLUSTER, ... | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5935 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
7C65
 
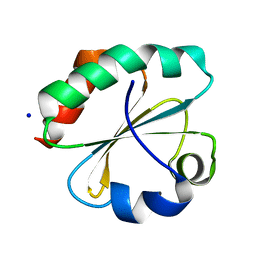 | | Crystal structure of thioredoxin m1 | | Descriptor: | SODIUM ION, Thioredoxin M1, chloroplastic | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-05-21 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
7C3F
 
 | | Crystal structure of ferredoxin: thioredoxin reductase and thioredoxin m2 complex | | Descriptor: | Ferredoxin-thioredoxin reductase catalytic chain, chloroplastic, Ferredoxin-thioredoxin reductase variable chain, ... | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3986 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
