1VD2
 
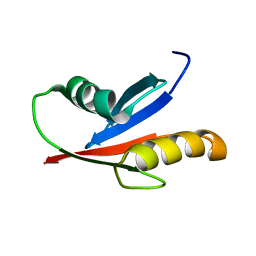 | | Solution Structure of the PB1 domain of PKCiota | | Descriptor: | Protein kinase C, iota type | | Authors: | Hirano, Y, Yoshinaga, S, Yokochi, M, Ogura, K, Noda, Y, Sumimoto, H, Inagaki, F. | | Deposit date: | 2004-03-18 | | Release date: | 2004-09-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of atypical protein kinase C PB1 domain and its mode of interaction with ZIP/p62 and MEK5
J.Biol.Chem., 279, 2004
|
|
1WMH
 
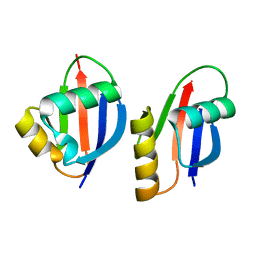 | | Crystal structure of a PB1 domain complex of Protein kinase c iota and Par6 alpha | | Descriptor: | Partitioning defective-6 homolog alpha, Protein kinase C, iota type | | Authors: | Hirano, Y, Yoshinaga, S, Suzuki, N.N, Horiuchi, M, Kohjima, M, Takeya, R, Sumimoto, H, Inagaki, F. | | Deposit date: | 2004-07-09 | | Release date: | 2004-12-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a Cell Polarity Regulator, a Complex between Atypical PKC and Par6 PB1 Domains
J.Biol.Chem., 280, 2005
|
|
9VBG
 
 | | Cryo-EM structure of human PLD3 apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Hirano, Y, Ezaki, W, Ohto, U, Shimizu, T. | | Deposit date: | 2025-06-04 | | Release date: | 2025-12-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanistic insights into single-stranded DNA degradation by lysosomal exonucleases PLD3 and PLD4 from structural snapshots.
Nat Commun, 2025
|
|
9VBH
 
 | | Cryo-EM structure of human PLD3 bound to ssDNA (poly(T)) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, DNA (5'-D(*TP*TP*TP*T)-3'), ... | | Authors: | Hirano, Y, Ezaki, W, Ohto, U, Shimizu, T. | | Deposit date: | 2025-06-04 | | Release date: | 2025-12-24 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Mechanistic insights into single-stranded DNA degradation by lysosomal exonucleases PLD3 and PLD4 from structural snapshots.
Nat Commun, 2025
|
|
9VBI
 
 | | Cryo-EM structure of PLD3 bound to ssDNA (poly(A)) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, DNA (5'-D(*AP*AP*AP*AP*A)-3'), ... | | Authors: | Hirano, Y, Ezaki, W, Ohto, U, Shimizu, T. | | Deposit date: | 2025-06-04 | | Release date: | 2025-12-24 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Mechanistic insights into single-stranded DNA degradation by lysosomal exonucleases PLD3 and PLD4 from structural snapshots.
Nat Commun, 2025
|
|
9VBK
 
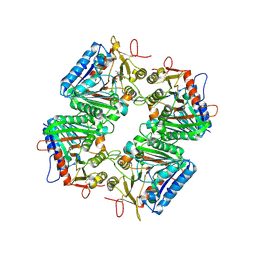 | | Cryo-EM structure of human PLD4 bound to ssDNA (poly(T)) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD4, ... | | Authors: | Hirano, Y, Ezaki, W, Ohto, U, Shimizu, T. | | Deposit date: | 2025-06-04 | | Release date: | 2025-12-24 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Mechanistic insights into single-stranded DNA degradation by lysosomal exonucleases PLD3 and PLD4 from structural snapshots.
Nat Commun, 2025
|
|
3VRC
 
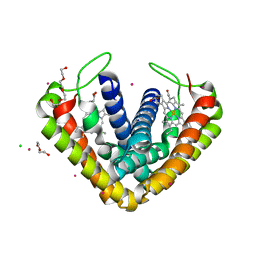 | | Crystal structure of cytochrome c' from Thermochromatium tepidum | | Descriptor: | CADMIUM ION, CHLORIDE ION, Cytochrome c', ... | | Authors: | Hirano, Y, Kimura, Y, Suzuki, H, Miki, K, Wang, Z.-Y. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure Analysis and Comparative Characterization of the Cytochrome c' and Flavocytochrome c from Thermophilic Purple Photosynthetic Bacterium Thermochromatium tepidum
Biochemistry, 51, 2012
|
|
3VRD
 
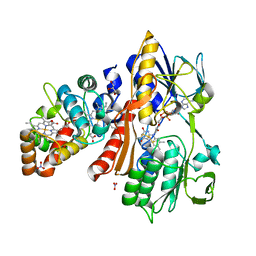 | | Crystal structure of flavocytochrome c from Thermochromatium tepidum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavocytochrome c flavin subunit, Flavocytochrome c heme subunit, ... | | Authors: | Hirano, Y, Kimura, Y, Suzuki, H, Miki, K, Wang, Z.-Y. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure Analysis and Comparative Characterization of the Cytochrome c' and Flavocytochrome c from Thermophilic Purple Photosynthetic Bacterium Thermochromatium tepidum
Biochemistry, 51, 2012
|
|
5Y04
 
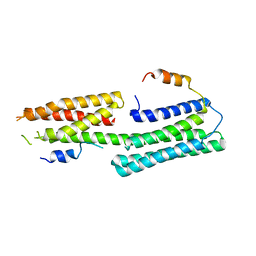 | |
9VBJ
 
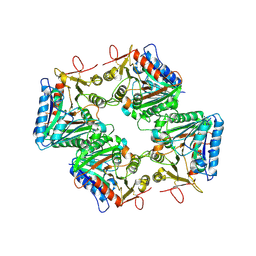 | | Cryo-EM structure of human PLD4 apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD4 | | Authors: | Hirano, Y, Ezaki, W, Ohto, U, Shimizu, T. | | Deposit date: | 2025-06-04 | | Release date: | 2025-12-24 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Mechanistic insights into single-stranded DNA degradation by lysosomal exonucleases PLD3 and PLD4 from structural snapshots.
Nat Commun, 2025
|
|
2Z4I
 
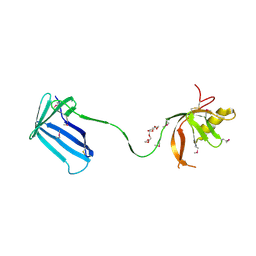 | | Crystal structure of the Cpx pathway activator NlpE from Escherichia coli | | Descriptor: | Copper homeostasis protein cutF, HEXAETHYLENE GLYCOL, SULFATE ION | | Authors: | Hirano, Y, Hossain, M.M, Takeda, K, Tokuda, H, Miki, K. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies of the Cpx Pathway Activator NlpE on the Outer Membrane of Escherichia coli
Structure, 15, 2007
|
|
2Z4H
 
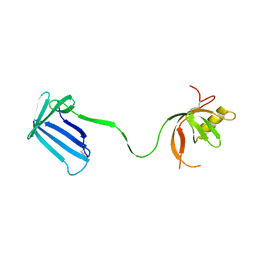 | | Crystal structure of the Cpx pathway activator NlpE from Escherichia coli | | Descriptor: | Copper homeostasis protein cutF, SULFATE ION | | Authors: | Hirano, Y, Hossain, M.M, Takeda, K, Tokuda, H, Miki, K. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of the Cpx Pathway Activator NlpE on the Outer Membrane of Escherichia coli
Structure, 15, 2007
|
|
9V6B
 
 | | Neutron crystal structure of the oxidized form of b5R at pD 6.5 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADH-cytochrome b5 reductase 3 | | Authors: | Hirano, Y, Kurihara, K, Kusaka, K, Ostermann, A, Hikita, M, Kimura, S, Miki, K, Tamada, T. | | Deposit date: | 2025-05-27 | | Release date: | 2025-11-19 | | Method: | NEUTRON DIFFRACTION (1.6 Å) | | Cite: | Structural basis of hydride and proton transfer reactions revealed by the detection of hydrogen atoms in mammalian NADH-cytochrome b 5 reductase.
Structure, 2025
|
|
9V6A
 
 | | X-ray crystal structure of the T66V mutant b5R co-crystallized with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3 | | Authors: | Hirano, Y, Kurihara, K, Kusaka, K, Ostermann, A, Hikita, M, Kimura, S, Miki, K, Tamada, T. | | Deposit date: | 2025-05-27 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Structural basis of hydride and proton transfer reactions revealed by the detection of hydrogen atoms in mammalian NADH-cytochrome b 5 reductase.
Structure, 2025
|
|
9V69
 
 | | X-ray crystal structure of the two-electron reduced form of wild type b5R | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hirano, Y, Kurihara, K, Kusaka, K, Ostermann, A, Hikita, M, Kimura, S, Miki, K, Tamada, T. | | Deposit date: | 2025-05-27 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Structural basis of hydride and proton transfer reactions revealed by the detection of hydrogen atoms in mammalian NADH-cytochrome b 5 reductase.
Structure, 2025
|
|
9V6C
 
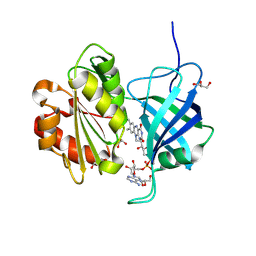 | | Neutron crystal structure of the oxidized form of b5R at pD 7.5 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADH-cytochrome b5 reductase 3 | | Authors: | Hirano, Y, Kurihara, K, Kusaka, K, Ostermann, A, Hikita, M, Kimura, S, Miki, K, Tamada, T. | | Deposit date: | 2025-05-27 | | Release date: | 2025-11-19 | | Method: | NEUTRON DIFFRACTION (1.45 Å) | | Cite: | Structural basis of hydride and proton transfer reactions revealed by the detection of hydrogen atoms in mammalian NADH-cytochrome b 5 reductase.
Structure, 2025
|
|
5XFL
 
 | |
5B3H
 
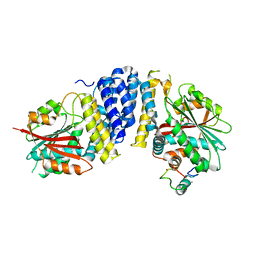 | | The crystal structure of the JACKDAW/IDD10 bound to the heterodimeric SHR-SCR complex | | Descriptor: | Protein SCARECROW, Protein SHORT-ROOT, ZINC ION, ... | | Authors: | Hirano, Y, Suyama, T, Nakagawa, M, Hakoshima, T. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD
Nat Plants, 3, 2017
|
|
8KCX
 
 | | Cryo-EM structure of human SIDT1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 1, ZINC ION, ... | | Authors: | Hirano, Y, Ohto, U, Shimizu, T. | | Deposit date: | 2023-08-08 | | Release date: | 2024-06-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM analysis reveals human SID-1 transmembrane family member 1 dynamics underlying lipid hydrolytic activity.
Commun Biol, 7, 2024
|
|
8KCW
 
 | | Cryo-EM structure of human SIDT1 bound to cholesterol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, SID1 transmembrane family member 1, ... | | Authors: | Hirano, Y, Ohto, U, Shimizu, T. | | Deposit date: | 2023-08-08 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Cryo-EM analysis reveals human SID-1 transmembrane family member 1 dynamics underlying lipid hydrolytic activity.
Commun Biol, 7, 2024
|
|
5D8V
 
 | | Ultra-high resolution structure of high-potential iron-sulfur protein | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Hirano, Y, Takeda, K, Miki, K. | | Deposit date: | 2015-08-18 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.48 Å) | | Cite: | Charge-density analysis of an iron-sulfur protein at an ultra-high resolution of 0.48 angstrom
Nature, 534, 2016
|
|
5B3G
 
 | | The crystal structure of the heterodimer of SHORT-ROOT and SCARECROW GRAS domains | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Hirano, Y, Nakagawa, M, Hakoshima, T. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD
Nat Plants, 3, 2017
|
|
3A9F
 
 | | Crystal structure of the C-terminal domain of cytochrome cz from Chlorobium tepidum | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Cytochrome c, HEME C, ... | | Authors: | Hirano, Y, Higuchi, M, Azai, C, Oh-oka, H, Miki, K, Wang, Z.-Y. | | Deposit date: | 2009-10-25 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the electron carrier domain of the reaction center cytochrome c(z) subunit from green photosynthetic bacterium Chlorobium tepidum
J.Mol.Biol., 397, 2010
|
|
3AU5
 
 | |
3AU4
 
 | |
