5YJX
 
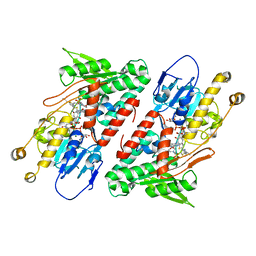 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with myxothiazol. | | Descriptor: | (2Z,6E)-7-{2'-[(2E,4E)-1,6-DIMETHYLHEPTA-2,4-DIENYL]-2,4'-BI-1,3-THIAZOL-4-YL}-3,5-DIMETHOXY-4-METHYLHEPTA-2,6-DIENAMID E, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5YJY
 
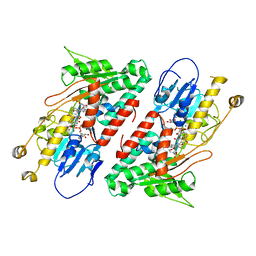 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with AC0-12. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-dodecyl-1-oxidanidyl-quinolin-1-ium-4-ol, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5YJW
 
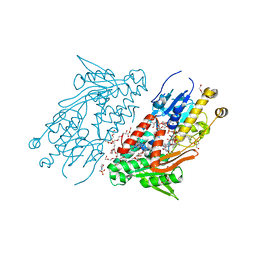 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with the competitive inhibitor, stigmatellin. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
8K5W
 
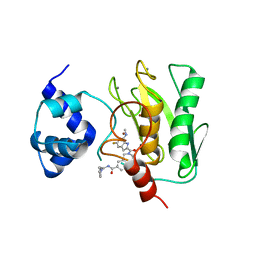 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | 2-[[5-fluoranyl-7-(methylamino)-1H-indol-2-yl]carbonyl]-N-(2-pyrrol-1-ylethyl)-3,4-dihydro-1H-isoquinoline-7-carboxamide, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5Y
 
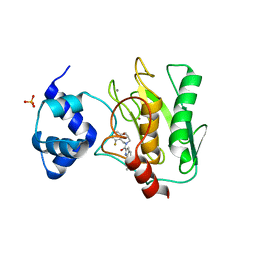 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | (3-azanyl-4-fluoranyl-5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)-[6-(2-oxidanylpropan-2-yl)-1H-indol-2-yl]methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5V
 
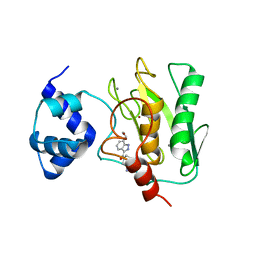 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | 6,7-dihydro-4H-[1,3]oxazolo[4,5-c]pyridin-5-yl-(7-ethyl-2H-indazol-3-yl)methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5X
 
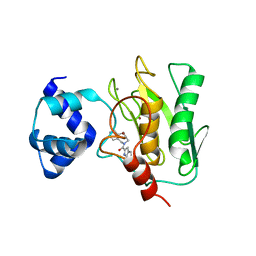 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | (6-cyclopropyl-1~{H}-indol-2-yl)-(5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)methanone, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
6M0D
 
 | | Beijerinckia indica beta-fructosyltransferase | | Descriptor: | Levansucrase, MAGNESIUM ION | | Authors: | Tonozuka, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 68 beta-fructosyltransferase from Beijerinckia indica subsp. indica in complex with fructose.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6M0E
 
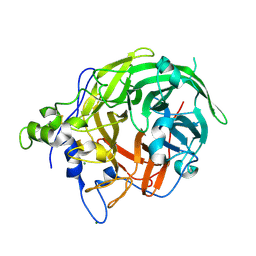 | | Beijerinckia indica beta-fructosyltransferase complexed with fructose | | Descriptor: | Levansucrase, MAGNESIUM ION, beta-D-fructofuranose, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 68 beta-fructosyltransferase from Beijerinckia indica subsp. indica in complex with fructose.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
5Y6Q
 
 | |
5XHA
 
 | |
5XH8
 
 | |
5XH9
 
 | | Aspergillus kawachii beta-fructofuranosidase | | Descriptor: | Extracellular invertase, SODIUM ION | | Authors: | Nagaya, M, Tonozuka, T. | | Deposit date: | 2017-04-19 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a beta-fructofuranosidase with high transfructosylation activity from Aspergillus kawachii
Biosci. Biotechnol. Biochem., 81, 2017
|
|
