1USW
 
 | | Crystal Structure of Ferulic Acid Esterase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FERULOYL ESTERASE A, SULFATE ION | | Authors: | Hermoso, J, Sanz-Aparicio, J, Molina, R, Faulds, C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-04-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of Feruloyl Esterase a from Aspergillus Niger Suggests Evolutive Functional Convergence in Feruloyl Esterase Family
J.Mol.Biol., 338, 2004
|
|
1ETH
 
 | | TRIACYLGLYCEROL LIPASE/COLIPASE COMPLEX | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Hermoso, J, Pignol, D, Kerfelec, B, Crenon, I, Chapus, C, Fontecilla-Camps, J.C. | | Deposit date: | 1995-09-13 | | Release date: | 1996-12-07 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lipase activation by nonionic detergents. The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether complex.
J.Biol.Chem., 271, 1996
|
|
2V05
 
 | |
1ZAE
 
 | | Solution structure of the functional domain of phi29 replication organizer p16.7c | | Descriptor: | Early protein GP16.7 | | Authors: | Asensio, J.L, Albert, A, Munoz-Espin, D, Gonzalez, C, Hermoso, J, Villar, L, Jimenez-Barbero, J, Salas, M, Meijer, W.J.J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the functional domain of phi29 replication organizer: insights into oligomerization and dna binding
J.Biol.Chem., 280, 2005
|
|
1AZD
 
 | | CONCANAVALIN FROM CANAVALIA BRASILIENSIS | | Descriptor: | CALCIUM ION, CONBR, MANGANESE (II) ION | | Authors: | Sanz-Aparicio, J, Hermoso, J, Grangeiro, T.B, Calvete, J.J, Cavada, B.S. | | Deposit date: | 1997-11-16 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of Canavalia brasiliensis lectin suggests a correlation between its quaternary conformation and its distinct biological properties from Concanavalin A.
FEBS Lett., 405, 1997
|
|
1GJR
 
 | | Ferredoxin-NADP+ Reductase complexed with NADP+ by COCRYSTALLIZATION | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-08-01 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of Coenzyme Recognition and Binding Revealed by Crystal Structure Analysis of Ferredoxin-Nadp(+) Reductase Complexed with Nadp(+)
J.Mol.Biol., 319, 2002
|
|
2V04
 
 | | CRYSTAL STRUCTURE OF CHOLINE BINDING PROTEIN F FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CHOLINE BINDING PROTEIN F, CHOLINE ION | | Authors: | Hermoso, J, Molina, R, Kahn, R, Stelter, M. | | Deposit date: | 2007-05-08 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cbpf, a Bifunctional Choline-Binding Protein and Autolysis Regulator from Streptococcus Pneumoniae.
Embo Rep., 10, 2009
|
|
1H85
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH VAL 136 REPLACED BY LEU (V136L) | | Descriptor: | FERREDOXIN--NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a Cluster of Hydrophobic Residues Near the Fad Cofactor in Anabaena Pcc 7119 Ferredoxin-Nadp+ Reductase for Optimal Complex Formation and Electron Transfer to Ferredoxin
J.Biol.Chem., 276, 2001
|
|
2W5U
 
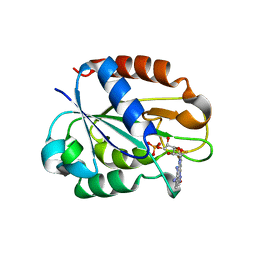 | | Flavodoxin from Helicobacter pylori in complex with the C3 inhibitor | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, [2-(5-amino-4-cyano-1H-pyrazol-1-yl)-5-(trifluoromethyl)phenyl](hydroxy)oxoammonium | | Authors: | Cremades, N, Perez-Dorado, I, Hermoso, J.A, Martinez-Julvez, M, Sancho, J. | | Deposit date: | 2008-12-12 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery of Specific Flavodoxin Inhibitors as Potential Therapeutic Agents Against Helicobacter Pylori Infection.
Acs Chem.Biol., 4, 2009
|
|
1BGG
 
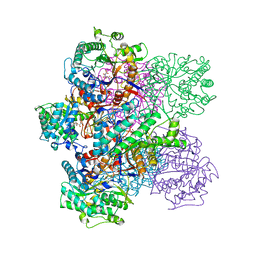 | | GLUCOSIDASE A FROM BACILLUS POLYMYXA COMPLEXED WITH GLUCONATE | | Descriptor: | BETA-GLUCOSIDASE A, D-gluconic acid | | Authors: | Sanz-Aparicio, J, Hermoso, J, Martinez-Ripoll, M, Polaina, J. | | Deposit date: | 1997-05-12 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of beta-glucosidase A from Bacillus polymyxa: insights into the catalytic activity in family 1 glycosyl hydrolases.
J.Mol.Biol., 275, 1998
|
|
3ESZ
 
 | | K2AK3A Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
1BQE
 
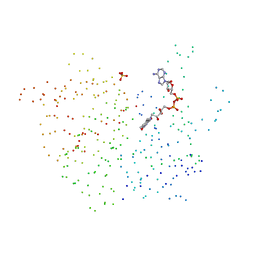 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH THR 155 REPLACED BY GLY (T155G) | | Descriptor: | FERREDOXIN--NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-08-14 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Probing the determinants of coenzyme specificity in ferredoxin-NADP+ reductase by site-directed mutagenesis.
J.Biol.Chem., 276, 2001
|
|
1BJK
 
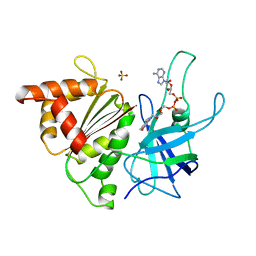 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH ARG 264 REPLACED BY GLU (R264E) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Arg100 and Arg264 from Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal NADP+ binding and electron transfer.
Biochemistry, 37, 1998
|
|
8A42
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AmiA in complex with an unknown peptide | | Descriptor: | Oligopeptide-binding protein AmiA, Unknown peptide | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2022-06-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLH
 
 | |
8QLG
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AliD in closed conformation in complex with Peptide 1 | | Descriptor: | AliD, Peptide 1, ZINC ION | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLJ
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with an unknown peptide | | Descriptor: | ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, Oligopeptide-binding protein AliB | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLC
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AliD in open conformation | | Descriptor: | AliD, MAGNESIUM ION | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLV
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with Peptide 4 | | Descriptor: | Oligopeptide-binding protein AliB, VAL-MET-VAL-LYS-GLY-PRO-GLY-PRO-GLY-ARG | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLK
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with Peptide 2 | | Descriptor: | ALA-ILE-GLN-SER-GLU-LYS-ALA-ARG-LYS-HIS-ASN, Oligopeptide-binding protein AliB | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLM
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with Peptide 3 | | Descriptor: | Oligopeptide-binding protein AliB, PRO-ILE-VAL-GLY-GLY-HIS-GLU-GLY-ALA-GLY-VAL | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QM0
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AmiA in complex with Peptide 5 | | Descriptor: | ALA-LYS-THR-ILE-LYS-ILE-THR-GLN-THR-ARG, Oligopeptide-binding protein AmiA | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
6RA9
 
 | | Novel structural features and post-translational modifications in eukaryotic elongation factor 1A2 from Oryctolagus cuniculus | | Descriptor: | Elongation factor 1-alpha 2, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Carriles, A.A, Hermoso, J, Gago, F. | | Deposit date: | 2019-04-05 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Cues for Understanding eEF1A2 Moonlighting.
Chembiochem, 22, 2021
|
|
1FJ0
 
 | |
1I8O
 
 | |
