1V58
 
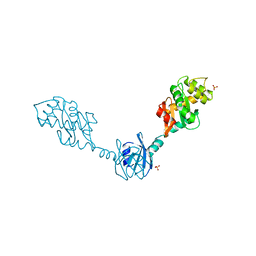 | | Crystal Structure Of the Reduced Protein Disulfide Bond Isomerase DsbG | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Heras, B, Edeling, M.A, Schirra, H.J, Raina, S, Martin, J.L. | | Deposit date: | 2003-11-21 | | Release date: | 2004-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the DsbG disulfide isomerase reveal an unstable disulfide
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1V57
 
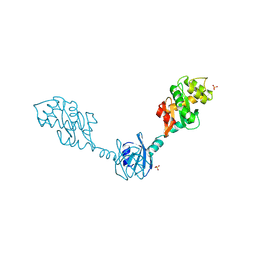 | | Crystal Structure of the Disulfide Bond Isomerase DsbG | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Heras, B, Edeling, M.A, Schirra, H.J, Raina, S, Martin, J.L. | | Deposit date: | 2003-11-21 | | Release date: | 2004-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the DsbG disulfide isomerase reveal an unstable disulfide
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3BCI
 
 | |
3L9U
 
 | |
3L9V
 
 | | Crystal Structure of Salmonella enterica serovar Typhimurium SrgA | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, HEXAETHYLENE GLYCOL, O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, ... | | Authors: | Heras, B, Jarrott, R, Shouldice, S.R, Guncar, G. | | Deposit date: | 2010-01-05 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural and functional characterization of three DsbA paralogues from Salmonella enterica serovar typhimurium
J.Biol.Chem., 285, 2010
|
|
3L9S
 
 | |
3BD2
 
 | |
3BCK
 
 | |
4KH3
 
 | | Structure of a bacterial self-associating protein | | Descriptor: | Antigen 43, MALONATE ION | | Authors: | Heras, B, Gee, C.L, Schembri, M.A, Totsika, M. | | Deposit date: | 2013-04-30 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6BQX
 
 | | Crystal structure of Escherichia coli DsbA in complex with N-methyl-1-(4-phenoxyphenyl)methanamine | | Descriptor: | N-methyl-1-(4-phenoxyphenyl)methanamine, Thiol:disulfide interchange protein DsbA | | Authors: | Heras, B, Totsika, M, Paxman, J.J, Wang, G, Scanlon, M.J. | | Deposit date: | 2017-11-29 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Inhibition of Diverse DsbA Enzymes in Multi-DsbA Encoding Pathogens.
Antioxid. Redox Signal., 29, 2018
|
|
6DNU
 
 | |
6DPS
 
 | |
6BR4
 
 | | Crystal structure of Escherichia coli DsbA in complex with {N}-methyl-1-(3-thiophen-2-ylphenyl)methanamine | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, ~{N}-methyl-1-(3-thiophen-2-ylphenyl)methanamine | | Authors: | Heras, B, Totsika, M, Paxman, J.J, Wang, G, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2017-11-29 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Inhibition of Diverse DsbA Enzymes in Multi-DsbA Encoding Pathogens.
Antioxid. Redox Signal., 29, 2018
|
|
5EK5
 
 | | STRUCTURAL CHARACTERIZATION OF IRMA FROM ESCHERICHIA COLI | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Heras, B, Moriel, D.G, Paxman, J.J, Schembri, M.A. | | Deposit date: | 2015-11-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Molecular and Structural Characterization of a Novel Escherichia coli Interleukin Receptor Mimic Protein.
Mbio, 7, 2016
|
|
7S1D
 
 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001877 (compound 39) | | Descriptor: | 1-[3-(thiophen-3-yl)benzyl]piperidin-2-one, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Caria, S. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
7S1F
 
 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001886 (compound 38) | | Descriptor: | 1-[(3-thiophen-3-ylphenyl)methyl]-3~{H}-pyrrol-2-one, COPPER (II) ION, GLYCEROL, ... | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Caria, S. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
7S1C
 
 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001897 (compound 1) | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, ~{N}-methyl-1-(3-thiophen-3-ylphenyl)methanamine | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Sharma, P. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
7S1L
 
 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001896 (compound 72) | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, methyl cis-4-({[3-(thiophen-3-yl)benzyl]amino}methyl)cyclohexanecarboxylate | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Caria, S. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
6PBU
 
 | | ClpP1 from Mycobacterium smegmatis | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, MALONATE ION | | Authors: | Heras, B, Nagpal, J, Dougan, D.A. | | Deposit date: | 2019-06-14 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular and structural insights into an asymmetric proteolytic complex (ClpP1P2) from Mycobacterium smegmatis.
Sci Rep, 9, 2019
|
|
9EDL
 
 | | Crystal structure of Francisella tularensis DsbA1 | | Descriptor: | Conserved hypothetical lipoprotein, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Penning, S, Heras, B, Paxman, J.J. | | Deposit date: | 2024-11-17 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Unveiling the versatility of the thioredoxin framework: Insights from the structural examination of Francisella tularensis DsbA1.
Comput Struct Biotechnol J, 23, 2024
|
|
6WHD
 
 | | Crystal structure of E.coli DsbA in complex with diaryl ether analogue 2 | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, [4-(4-cyano-3-methylphenoxy)phenyl]acetic acid | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiLX).
J.Med.Chem., 63, 2020
|
|
7TTV
 
 | |
6BEA
 
 | |
3A3T
 
 | | The oxidoreductase NmDsbA1 from N. meningitidis | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Vivian, J.P, Scoullar, J, Bushell, S, Beddoe, T, Wilce, M.C.J, Byres, E, Boyle, T.P, Rimmer, K, Doak, B, Simpson, J.S, Graham, B, Heras, B, Kahler, C.M, Rossjohn, J, Scanlon, M.J. | | Deposit date: | 2009-06-19 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the oxidoreductase DsbA1 from Neisseria meningitidis
J.Mol.Biol., 394, 2009
|
|
3F4R
 
 | | Crystal structure of Wolbachia pipientis alpha-DsbA1 | | Descriptor: | PENTAETHYLENE GLYCOL, Putative uncharacterized protein, TRIETHYLENE GLYCOL | | Authors: | Kurz, M, Heras, B, Martin, J.L. | | Deposit date: | 2008-11-02 | | Release date: | 2009-03-24 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of the Oxidoreductase alpha-DsbA1 from Wolbachia pipientis.
ANTIOXID.REDOX SIGNAL., 11, 2009
|
|
