1OQA
 
 | | Solution structure of the BRCT-c domain from human BRCA1 | | Descriptor: | Breast cancer type 1 susceptibility protein | | Authors: | Gaiser, O.J, Ball, L.J, Schmieder, P, Leitner, D, Strauss, H, Wahl, M, Kuhne, R, Oschkinat, H, Heinemann, U. | | Deposit date: | 2003-03-07 | | Release date: | 2004-06-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics, and association behavior of the C-terminal BRCT domain from the breast cancer-associated protein BRCA1.
Biochemistry, 43, 2004
|
|
1PMJ
 
 | | Crystal structure of Caldicellulosiruptor saccharolyticus CBM27-1 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Roske, Y, Sunna, A, Heinemann, U. | | Deposit date: | 2003-06-11 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution crystal structures of Caldicellulosiruptor strain Rt8B.4 carbohydrate-binding module CBM27-1 and its complex with mannohexaose.
J.Mol.Biol., 340, 2004
|
|
1PMH
 
 | | Crystal structure of Caldicellulosiruptor saccharolyticus CBM27-1 in complex with mannohexaose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, beta-1,4-mannanase, ... | | Authors: | Roske, Y, Sunna, A, Heinemann, U. | | Deposit date: | 2003-06-11 | | Release date: | 2004-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | High-resolution crystal structures of Caldicellulosiruptor strain Rt8B.4 carbohydrate-binding module CBM27-1 and its complex with mannohexaose.
J.Mol.Biol., 340, 2004
|
|
1QYM
 
 | | X-ray structure of human gankyrin | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10 | | Authors: | Manjasetty, B.A, Quedenau, C, Sievert, V, Buessow, K, Niesen, F, Delbrueck, H, Heinemann, U. | | Deposit date: | 2003-09-11 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of human gankyrin, the product of a gene linked to hepatocellular carcinoma.
Proteins, 55, 2004
|
|
1R71
 
 | | Crystal Structure of the DNA binding domain of KorB in complex with the operator DNA | | Descriptor: | 5'-D(*AP*(BRU)P*TP*TP*TP*AP*GP*CP*GP*GP*CP*TP*AP*AP*AP*AP*G)-3', 5'-D(*CP*(BRU)P*TP*TP*TP*AP*GP*CP*CP*GP*CP*TP*AP*AP*AP*AP*(BRU))-3', Transcriptional repressor protein korB | | Authors: | Khare, D, Ziegelin, G, Lanka, E, Heinemann, U. | | Deposit date: | 2003-10-17 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SAW
 
 | | X-ray structure of homo sapiens protein FLJ36880 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, hypothetical protein FLJ36880 | | Authors: | Manjasetty, B.A, Niesen, F.H, Delbrueck, H, Goetz, F, Sievert, V, Buessow, K, Behlke, J, Heinemann, U. | | Deposit date: | 2004-02-09 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of fumarylacetoacetate hydrolase family member Homo sapiens FLJ36880.
Biol.Chem., 385, 2004
|
|
1SDR
 
 | | CRYSTAL STRUCTURE OF AN RNA DODECAMER CONTAINING THE ESCHERICHIA COLI SHINE-DALGARNO SEQUENCE | | Descriptor: | RNA (5'-R(*AP*UP*CP*AP*CP*CP*UP*CP*CP*UP*UP*A)-3'), RNA (5'-R(*UP*AP*AP*GP*GP*AP*GP*GP*UP*GP*AP*U)-3') | | Authors: | Schindelin, H, Zhang, M, Bald, R, Fuerste, J.-P, Erdmann, V.A, Heinemann, U. | | Deposit date: | 1994-12-11 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an RNA dodecamer containing the Escherichia coli Shine-Dalgarno sequence.
J.Mol.Biol., 249, 1995
|
|
6G5W
 
 | | Crystal Structure of KDM4A with compound YP-03-038 | | Descriptor: | (4~{R})-5-methyl-4-phenyl-2-pyridin-2-yl-pyrazolidin-3-one, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | Malecki, P.H, Carter, D.M, Gohlke, U, Specker, E, Nazare, M, Weiss, M.S, Heinemann, U. | | Deposit date: | 2018-03-30 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Enhanced Properties of a Benzimidazole Benzylpyrazole Lysine Demethylase Inhibitor: Mechanism-of-Action, Binding Site Analysis, and Activity in Cellular Models of Prostate Cancer.
J.Med.Chem., 64, 2021
|
|
6G5X
 
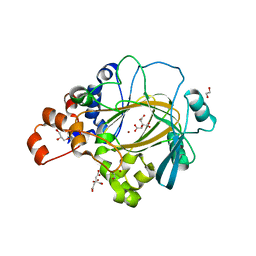 | | Crystal Structure of KDM4A with compound YP-02-145 | | Descriptor: | 1,2-ETHANEDIOL, 2-(3-methyl-5-oxidanylidene-4-phenyl-4~{H}-pyrazol-1-yl)-3~{H}-benzimidazole-5-carboxylic acid, CITRIC ACID, ... | | Authors: | Malecki, P.H, Carter, D.M, Gohlke, U, Specker, E, Nazare, M, Weiss, M.S, Heinemann, U. | | Deposit date: | 2018-03-30 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Enhanced Properties of a Benzimidazole Benzylpyrazole Lysine Demethylase Inhibitor: Mechanism-of-Action, Binding Site Analysis, and Activity in Cellular Models of Prostate Cancer.
J.Med.Chem., 64, 2021
|
|
1GPP
 
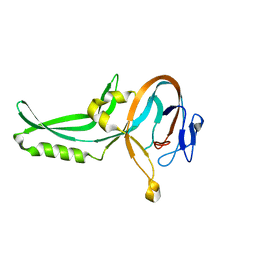 | |
1GZU
 
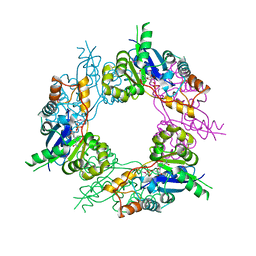 | | Crystal Structure of Human Nicotinamide Mononucleotide Adenylyltransferase in Complex with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE | | Authors: | Werner, E, Ziegler, M, Lerner, F, Schweiger, M, Heinemann, U. | | Deposit date: | 2002-06-06 | | Release date: | 2002-07-05 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human nicotinamide mononucleotide adenylyltransferase in complex with NMN.
FEBS Lett., 516, 2002
|
|
3ZM1
 
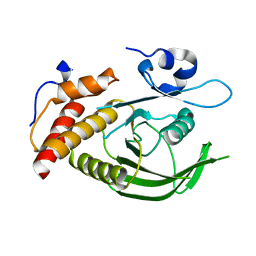 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
3ZM0
 
 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
3ZM3
 
 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
3ZM2
 
 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
4A4I
 
 | | Crystal structure of the human Lin28b cold shock domain | | Descriptor: | GLYCEROL, PROTEIN LIN-28 HOMOLOG B, SULFATE ION | | Authors: | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | Deposit date: | 2011-10-14 | | Release date: | 2012-08-08 | | Last modified: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Lin28 Cold-Shock Domain Remodels Pre-Let-7 Microrna.
Nucleic Acids Res., 40, 2012
|
|
4A76
 
 | | The Lin28b Cold shock domain in complex with heptathymidine | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP)-3', LIN28 COLD SHOCK DOMAIN | | Authors: | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | Deposit date: | 2011-11-11 | | Release date: | 2012-09-05 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The Lin28 Cold-Shock Domain Remodels Pre-Let-7 Microrna.
Nucleic Acids Res., 40, 2012
|
|
4A75
 
 | | The Lin28b Cold shock domain in complex with hexathymidine. | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP)-3', LIN28 COLD SHOCK DOMAIN, THIOCYANATE ION | | Authors: | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | Deposit date: | 2011-11-11 | | Release date: | 2012-09-05 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Lin28 Cold-Shock Domain Remodels Pre-Let-7 Microrna.
Nucleic Acids Res., 40, 2012
|
|
4ALP
 
 | | The Lin28b Cold shock domain in complex with hexauridine | | Descriptor: | GLYCEROL, HEXA URIDINE, LIN28 ISOFORM B | | Authors: | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | Deposit date: | 2012-03-05 | | Release date: | 2012-09-05 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The Lin28 Cold-Shock Domain Remodels Pre-Let-7 Microrna.
Nucleic Acids Res., 40, 2012
|
|
4AEN
 
 | | HLA-DR1 with covalently linked CLIP106-120 in reversed orientation | | Descriptor: | GLYCEROL, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN GAMMA CHAIN, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Schlundt, A, Guenther, S, Sticht, J, Wieczorek, M, Roske, Y, Heinemann, U, Freund, C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide Linkage to the Alpha-Subunit of Mhcii Creates a Stably Inverted Antigen Presentation Complex.
J.Mol.Biol., 423, 2012
|
|
4AH2
 
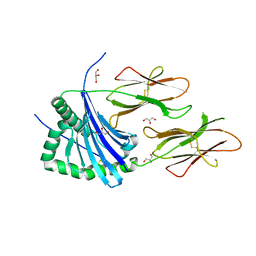 | | HLA-DR1 with covalently linked CLIP106-120 in canonical orientation | | Descriptor: | GLYCEROL, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN GAMMA CHAIN, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN,DRB1-1 BETA CHAIN, ... | | Authors: | Schlundt, A, Guenther, S, Sticht, J, Wieczorek, M, Roske, Y, Heinemann, U, Freund, C. | | Deposit date: | 2012-02-03 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Peptide Linkage to the Alpha-Subunit of Mhcii Creates a Stably Inverted Antigen Presentation Complex.
J.Mol.Biol., 423, 2012
|
|
2VTG
 
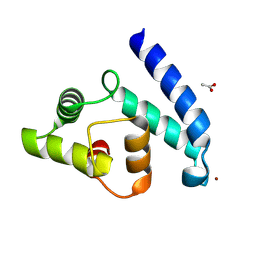 | | Crystal Structure of Human Iba2, trigonal crystal form | | Descriptor: | ACETATE ION, IONIZED CALCIUM-BINDING ADAPTER MOLECULE 2, ZINC ION | | Authors: | Schulze, J.O, Quedenau, C, Roske, Y, Turnbull, A, Mueller, U, Heinemann, U, Buessow, K. | | Deposit date: | 2008-05-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and Functional Characterization of Human Iba Proteins.
FEBS J., 275, 2008
|
|
1J75
 
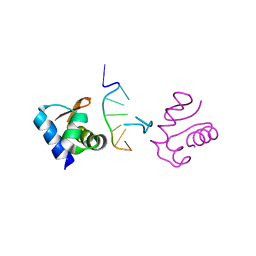 | | Crystal Structure of the DNA-Binding Domain Zalpha of DLM-1 Bound to Z-DNA | | Descriptor: | 5'-D(*TP*CP*GP*CP*GP*CP*G)-3', Tumor Stroma and Activated Macrophage Protein DLM-1 | | Authors: | Schwartz, T, Behlke, J, Lowenhaupt, K, Heinemann, U, Rich, A. | | Deposit date: | 2001-05-15 | | Release date: | 2001-09-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins.
Nat.Struct.Biol., 8, 2001
|
|
1IGQ
 
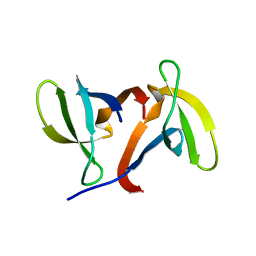 | |
1IGU
 
 | |
