2LKN
 
 | | Solution structure of the PPIase domain of human aryl-hydrocarbon receptor-interacting protein (AIP) | | Descriptor: | AH receptor-interacting protein | | Authors: | Linnert, M, Lin, Y, Manns, A, Haupt, K, Paschke, A, Fischer, G, Weiwad, M, Luecke, C. | | Deposit date: | 2011-10-17 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The FKBP-Type Domain of the Human Aryl Hydrocarbon Receptor-Interacting Protein Reveals an Unusual Hsp90 Interaction.
Biochemistry, 52, 2013
|
|
1J75
 
 | | Crystal Structure of the DNA-Binding Domain Zalpha of DLM-1 Bound to Z-DNA | | Descriptor: | 5'-D(*TP*CP*GP*CP*GP*CP*G)-3', Tumor Stroma and Activated Macrophage Protein DLM-1 | | Authors: | Schwartz, T, Behlke, J, Lowenhaupt, K, Heinemann, U, Rich, A. | | Deposit date: | 2001-05-15 | | Release date: | 2001-09-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins.
Nat.Struct.Biol., 8, 2001
|
|
1XMK
 
 | | The Crystal structure of the Zb domain from the RNA editing enzyme ADAR1 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Double-stranded RNA-specific adenosine deaminase, ... | | Authors: | Athanasiadis, A, Placido, D, Maas, S, Brown II, B.A, Lowenhaupt, K, Rich, A. | | Deposit date: | 2004-10-03 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The Crystal Structure of the Z[beta] Domain of the RNA-editing Enzyme ADAR1 Reveals Distinct Conserved Surfaces Among Z-domains.
J.Mol.Biol., 351, 2005
|
|
2ACJ
 
 | | Crystal structure of the B/Z junction containing DNA bound to Z-DNA binding proteins | | Descriptor: | 5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*GP*GP*CP*GP*CP*GP*CP*G)-3', 5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*CP*AP*TP*AP*AP*AP*CP*C)-3', Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Lowenhaupt, K, Rich, A, Kim, Y.-G, Kim, K.K. | | Deposit date: | 2005-07-19 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases.
Nature, 437, 2005
|
|
1QGP
 
 | | NMR STRUCTURE OF THE Z-ALPHA DOMAIN OF ADAR1, 15 STRUCTURES | | Descriptor: | PROTEIN (DOUBLE STRANDED RNA ADENOSINE DEAMINASE) | | Authors: | Schade, M, Turner, C.J, Kuehne, R, Schmieder, P, Lowenhaupt, K, Herbert, A, Rich, A, Oschkinat, H. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Zalpha domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1OYI
 
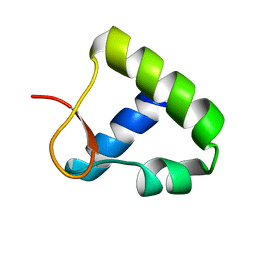 | | Solution structure of the Z-DNA binding domain of the vaccinia virus gene E3L | | Descriptor: | double-stranded RNA-binding protein | | Authors: | Kahmann, J.D, Wecking, D.A, Putter, V, Lowenhaupt, K, Kim, Y.-G, Schmieder, P, Oschkinat, H, Rich, A, Schade, M. | | Deposit date: | 2003-04-04 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of E3L shows a tyrosine conformation that may explain its reduced affinity to Z-DNA in vitro.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SFU
 
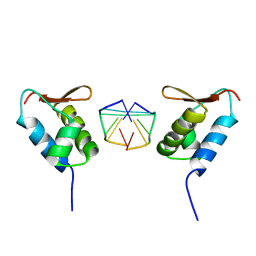 | | Crystal structure of the viral Zalpha domain bound to left-handed Z-DNA | | Descriptor: | 34L protein, 5'-D(*T*CP*GP*CP*GP*CP*G)-3' | | Authors: | Ha, S.C, Van Quyen, D, Wu, C.A, Lowenhaupt, K, Rich, A, Kim, Y.G, Kim, K.K. | | Deposit date: | 2004-02-20 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3EY6
 
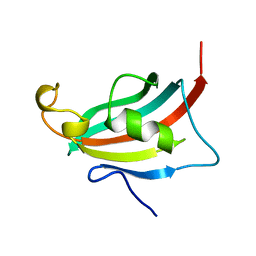 | | Crystal structure of the FK506-binding domain of human FKBP38 | | Descriptor: | FK506-binding protein 8 | | Authors: | Parthier, C, Maestre-Martinez, M, Neumann, P, Edlich, F, Fischer, G, Luecke, C, Stubbs, M.T. | | Deposit date: | 2008-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A charge-sensitive loop in the FKBP38 catalytic domain modulates Bcl-2 binding.
J.Mol.Recognit., 24, 2011
|
|
5EBI
 
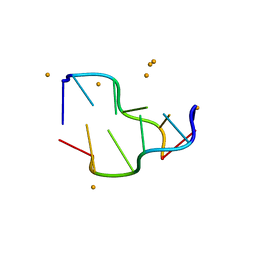 | | Crystal structure of a DNA-RNA chimera in complex with Ba2+ ions: a case of unusual multi-domain twinning | | Descriptor: | BARIUM ION, DNA/RNA (5'-D(*C)-R(P*G)-D(P*C)-R(P*G)-D(P*C)-R(P*G)-3') | | Authors: | Gilski, M, Drozdzal, P, Kierzek, R, Jaskolski, M. | | Deposit date: | 2015-10-19 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Atomic resolution structure of a chimeric DNA-RNA Z-type duplex in complex with Ba(2+) ions: a case of complicated multi-domain twinning.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2L54
 
 | | Solution structure of the Zalpha domain mutant of ADAR1 (N43A,Y47A) | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Zhao, J, Pervushin, K, Feng, S, Droge, P. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Alternate rRNA secondary structures as regulators of translation
Nat.Struct.Mol.Biol., 18, 2011
|
|
1QBJ
 
 | | CRYSTAL STRUCTURE OF THE ZALPHA Z-DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), PROTEIN (DOUBLE-STRANDED RNA SPECIFIC ADENOSINE DEAMINASE (ADAR1)) | | Authors: | Schwartz, T, Rould, M.A, Rich, A. | | Deposit date: | 1999-04-22 | | Release date: | 1999-07-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA.
Science, 284, 1999
|
|
2GXB
 
 | | Crystal Structure of The Za Domain bound to Z-RNA | | Descriptor: | 5'-R(P*(DU)P*CP*GP*CP*GP*CP*G)-3', Double-stranded RNA-specific adenosine deaminase, SODIUM ION | | Authors: | Athanasiadis, A, Placido, D, Rich, A. | | Deposit date: | 2006-05-08 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Left-Handed RNA Double Helix Bound by the Zalpha Domain of the RNA-Editing Enzyme ADAR1.
Structure, 15, 2007
|
|
