1G94
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE TERNARY COMPLEX BETWEEN PSYCHROPHILIC ALPHA AMYLASE FROM PSEUDOALTEROMONAS HALOPLANCTIS IN COMPLEX WITH A HEPTA-SACCHARIDE AND A TRIS MOLECULE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Aghajari, N, Roth, M, Haser, R. | | Deposit date: | 2000-11-22 | | Release date: | 2001-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic evidence of a transglycosylation reaction: ternary complexes of a psychrophilic alpha-amylase.
Biochemistry, 41, 2002
|
|
1AQM
 
 | | ALPHA-AMYLASE FROM ALTEROMONAS HALOPLANCTIS COMPLEXED WITH TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 1997-07-31 | | Release date: | 1999-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor.
Protein Sci., 7, 1998
|
|
1G9H
 
 | | TERNARY COMPLEX BETWEEN PSYCHROPHILIC ALPHA-AMYLASE, COMII (PSEUDO TRI-SACCHARIDE FROM BAYER) AND TRIS (2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose, ALPHA-AMYLASE, ... | | Authors: | Aghajari, N, Roth, M, Haser, R. | | Deposit date: | 2000-11-23 | | Release date: | 2002-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic evidence of a transglycosylation reaction: ternary complexes of a
psychrophilic alpha-amylase.
Biochemistry, 41, 2002
|
|
1AQH
 
 | | ALPHA-AMYLASE FROM ALTEROMONAS HALOPLANCTIS | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 1997-07-30 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor.
Protein Sci., 7, 1998
|
|
1AMY
 
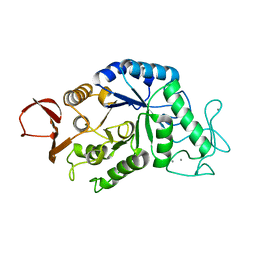 | |
1G9G
 
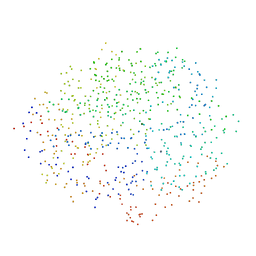 | | XTAL-STRUCTURE OF THE FREE NATIVE CELLULASE CEL48F | | Descriptor: | CALCIUM ION, CELLULASE CEL48F, MAGNESIUM ION | | Authors: | Parsiegla, G, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-11-23 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of mutants of cellulase Cel48F of Clostridium cellulolyticum in complex with long hemithiocellooligosaccharides give rise to a new view of the substrate pathway during processive action
J.Mol.Biol., 375, 2008
|
|
1G9J
 
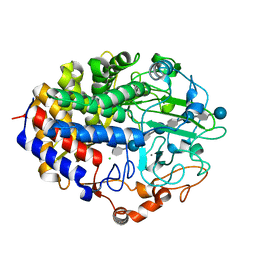 | | X-TAL STRUCTURE OF THE MUTANT E44Q OF THE CELLULASE CEL48F IN COMPLEX WITH A THIOOLIGOSACCHARIDE | | Descriptor: | CALCIUM ION, CELLULASE CEL48F, CHLORIDE ION, ... | | Authors: | Parsiegla, G, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-11-24 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of mutants of cellulase Cel48F of Clostridium cellulolyticum in complex with long hemithiocellooligosaccharides give rise to a new view of the substrate pathway during processive action
J.Mol.Biol., 375, 2008
|
|
1JD7
 
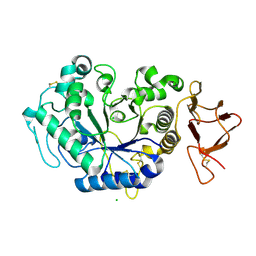 | |
1JD9
 
 | |
1TVN
 
 | |
1TVP
 
 | |
1BG9
 
 | | BARLEY ALPHA-AMYLASE WITH SUBSTRATE ANALOGUE ACARBOSE | | Descriptor: | 1,4-ALPHA-D-GLUCAN GLUCANOHYDROLASE, 4,6-dideoxy-4-{[(1S,4S,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Kadziola, A, Haser, R. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular structure of a barley alpha-amylase-inhibitor complex: implications for starch binding and catalysis.
J.Mol.Biol., 278, 1998
|
|
1FAE
 
 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellobiose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-13 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1F9O
 
 | | Crystal structure of the cellulase Cel48F from C. Cellulolyticum with the thiooligosaccharide inhibitor PIPS-IG3 | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-iodophenyl 1,4-dithio-beta-D-glucopyranoside | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-11 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1F9D
 
 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellotetraose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-10 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1FBW
 
 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellohexaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1AQE
 
 | |
1P6W
 
 | | Crystal structure of barley alpha-amylase isozyme 1 (AMY1) in complex with the substrate analogue, methyl 4I,4II,4III-tri-thiomaltotetraoside (thio-DP4) | | Descriptor: | CALCIUM ION, PROTEIN (Alpha-amylase type A isozyme), alpha-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose, ... | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2003-04-30 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of barley alpha-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs
Structure, 11, 2003
|
|
1B0I
 
 | | ALPHA-AMYLASE FROM ALTEROMONAS HALOPLANCTIS | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (ALPHA-AMYLASE) | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 1998-11-10 | | Release date: | 1999-11-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the psychrophilic Alteromonas haloplanctis alpha-amylase give insights into cold adaptation at a molecular level.
Structure, 6, 1998
|
|
1AVA
 
 | | AMY2/BASI PROTEIN-PROTEIN COMPLEX FROM BARLEY SEED | | Descriptor: | BARLEY ALPHA-AMYLASE 2(CV MENUET), BARLEY ALPHA-AMYLASE/SUBTILISIN INHIBITOR, CALCIUM ION | | Authors: | Vallee, F, Kadziola, A, Bourne, Y, Juy, M, Svensson, B, Haser, R. | | Deposit date: | 1997-09-15 | | Release date: | 1999-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Barley alpha-amylase bound to its endogenous protein inhibitor BASI: crystal structure of the complex at 1.9 A resolution.
Structure, 6, 1998
|
|
1FBO
 
 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellobiitol | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-D-glucose | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-16 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1CZJ
 
 | | CYTOCHROME C OF CLASS III (AMBLER) 26 KD | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Czjzek, M, Haser, R. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of a dimeric octaheme cytochrome c3 (M(r) 26,000) from Desulfovibrio desulfuricans Norway.
Structure, 4, 1996
|
|
1DKH
 
 | | CRYSTAL STRUCTURE OF THE HEMOPHORE HASA, PH 6.5 | | Descriptor: | HEME-BINDING PROTEIN A, PROTOPORPHYRIN IX CONTAINING FE, SAMARIUM (III) ION, ... | | Authors: | Arnoux, P, Haser, R, Izadi-Pruneyre, N, Lecroisey, A, Czjzek, M. | | Deposit date: | 1999-12-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Functional aspects of the heme bound hemophore HasA by structural analysis of various crystal forms.
Proteins, 41, 2000
|
|
1HT6
 
 | | CRYSTAL STRUCTURE AT 1.5A RESOLUTION OF THE BARLEY ALPHA-AMYLASE ISOZYME 1 | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-AMYLASE ISOZYME 1, CALCIUM ION | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2000-12-29 | | Release date: | 2003-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of barley alpha-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs
Structure, 11, 2003
|
|
1DK0
 
 | | CRYSTAL STRUCTURE OF THE HEMOPHORE HASA FROM SERRATIA MARCESCENS CRYSTAL FORM P2(1), PH8 | | Descriptor: | HEME-BINDING PROTEIN A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnoux, P, Haser, R, Izadi-Pruneyre, N, Lecroisey, A, Czjzek, M. | | Deposit date: | 1999-12-06 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Functional aspects of the heme bound hemophore HasA by structural analysis of various crystal forms.
Proteins, 41, 2000
|
|
