9CLB
 
 | | Crystal structure of Bak bound to the inhibitory aBAK | | Descriptor: | Bcl-2 homologous antagonist/killer, aBAK | | Authors: | Birkinshaw, R.W, Berger, S.A, Lee, E.F, Harris, T.J, Tran, S, Bera, A.K, Arguinchona, L, Kang, A, Sankaran, B, Kasapgil, S, Miller, M.S, Smyth, S, Uren, R, Kluck, R, Colman, P.M, Fairlie, W.D, Czabotar, P.E, Baker, D. | | Deposit date: | 2024-07-10 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Computational design of potent and selective inhibitors of BAK and BAX
To Be Published
|
|
1O65
 
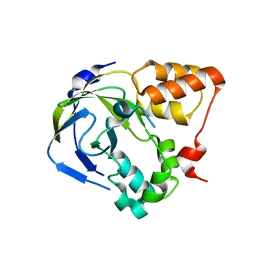 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yiiM | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O62
 
 | |
6V4M
 
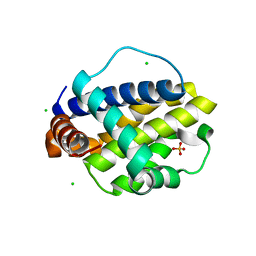 | | Trichuris suis BCL-2 | | Descriptor: | BCL-2, CHLORIDE ION, SULFATE ION | | Authors: | Fairlie, W.D, Lee, E.F, Smith, B.J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Diversity in the intrinsic apoptosis pathway of nematodes.
Commun Biol, 3, 2020
|
|
6DCN
 
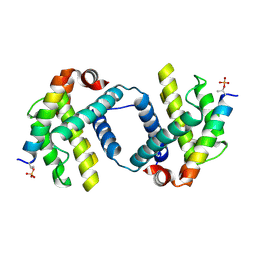 | | Bcl-xL complex with Beclin 1 BH3 domain T108pThr | | Descriptor: | BCL-xl protein, Beclin-1 | | Authors: | Lee, E.F, Smith, B.J, Smith, N.A, Yao, S, Fairlie, W.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Autophagy, 15, 2019
|
|
6DCO
 
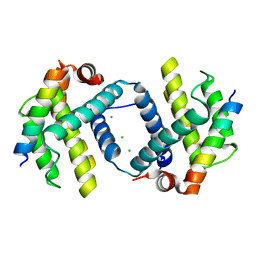 | | Bcl-xL complex with Beclin 1 BH3 domain T108D | | Descriptor: | Bcl-2-like protein 1,Bcl-2-like protein 1, Beclin-1, CHLORIDE ION | | Authors: | Lee, E.F, Smith, B.J, Smith, N.A, Yao, S, Fairlie, W.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Autophagy, 15, 2019
|
|
2TMP
 
 | | N-TERMINAL DOMAIN OF TISSUE INHIBITOR OF METALLOPROTEINASE-2 (N-TIMP-2), NMR, 49 STRUCTURES | | Descriptor: | TISSUE INHIBITOR OF METALLOPROTEINASES-2 | | Authors: | Muskett, F.W, Frenkiel, T.A, Feeney, J, Freedman, R.B, Carr, M.D, Williamson, R.A. | | Deposit date: | 1998-05-26 | | Release date: | 1998-12-09 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | High resolution structure of the N-terminal domain of tissue inhibitor of metalloproteinases-2 and characterization of its interaction site with matrix metalloproteinase-3.
J.Biol.Chem., 273, 1998
|
|
7JGV
 
 | | CRYSTAL STRUCTURE OF BCL-XL IN COMPLEX WITH COMPOUND 1620116, CRYSTAL FORM 2 | | Descriptor: | 6-[(8E)-8-{2-[4-(benzylcarbamoyl)-1,3-thiazol-2-yl]hydrazinylidene}-5,6,7,8-tetrahydronaphthalen-2-yl]-3-(2-phenylethoxy)pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Lee, M, Fairlie, W.D, Smith, B.J, Lee, E.F. | | Deposit date: | 2020-07-19 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
7JMT
 
 | | Crystal structure of schistosome BCL-2 bound to ABT-737 | | Descriptor: | 4-{4-[(4'-CHLOROBIPHENYL-2-YL)METHYL]PIPERAZIN-1-YL}-N-{[4-({(1R)-3-(DIMETHYLAMINO)-1-[(PHENYLTHIO)METHYL]PROPYL}AMINO)-3-NITROPHENYL]SULFONYL}BENZAMIDE, BCL-2 protein | | Authors: | Smith, N.A, Smith, B.J, Lee, E.F, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2020-08-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
7JGW
 
 | | Crystal structure of BCL-XL in complex with COMPOUND 1620116, CRYSTAL FORM 1 | | Descriptor: | 6-[(8E)-8-{2-[4-(benzylcarbamoyl)-1,3-thiazol-2-yl]hydrazinylidene}-5,6,7,8-tetrahydronaphthalen-2-yl]-3-(2-phenylethoxy)pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Lee, M, Fairlie, W.D, Smith, B.J, Lee, E.F. | | Deposit date: | 2020-07-19 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
1O63
 
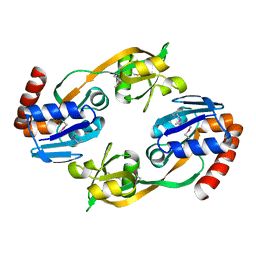 | |
1O6C
 
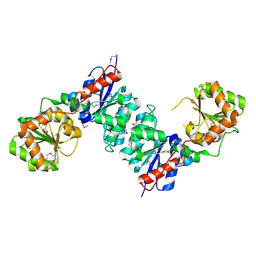 | |
1O6B
 
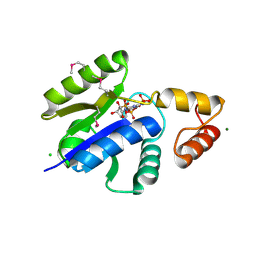 | |
1O66
 
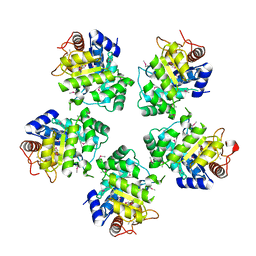 | |
1O67
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yiiM | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O60
 
 | | Crystal structure of KDO-8-phosphate synthase | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O61
 
 | | Crystal structure of a PLP-dependent enzyme with PLP | | Descriptor: | ACETATE ION, PYRIDOXAL-5'-PHOSPHATE, aminotransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O68
 
 | |
1O6D
 
 | | Crystal structure of a hypothetical protein | | Descriptor: | Hypothetical UPF0247 protein TM0844 | | Authors: | Structural GenomiX | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O64
 
 | |
1O69
 
 | | Crystal structure of a PLP-dependent enzyme | | Descriptor: | (2-AMINO-4-FORMYL-5-HYDROXY-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, BETA-MERCAPTOETHANOL, aminotransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1J6X
 
 | | CRYSTAL STRUCTURE OF HELICOBACTER PYLORI LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
1INN
 
 | | CRYSTAL STRUCTURE OF D. RADIODURANS LUXS, P21 | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
1J6W
 
 | | CRYSTAL STRUCTURE OF HAEMOPHILUS INFLUENZAE LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
1J6V
 
 | | CRYSTAL STRUCTURE OF D. RADIODURANS LUXS, C2 | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
