2DFB
 
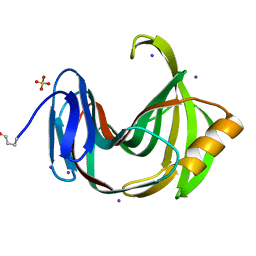 | | Xylanase II from Tricoderma reesei at 100K | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, SULFATE ION | | Authors: | Harata, K, Akiba, T. | | Deposit date: | 2006-02-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structure of an orthorhombic form of xylanase II from Trichoderma reesei and analysis of thermal displacement.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
1TAY
 
 | |
1TBY
 
 | |
1TDY
 
 | |
1TCY
 
 | |
1D6Q
 
 | | HUMAN LYSOZYME E102 MUTANT LABELLED WITH 2',3'-EPOXYPROPYL GLYCOSIDE OF N-ACETYLLACTOSAMINE | | Descriptor: | GLYCEROL, LYSOZYME, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1999-10-15 | | Release date: | 2000-01-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Protein-carbohydrate interactions in human lysozyme probed by combining site-directed mutagenesis and affinity labeling.
Biochemistry, 39, 2000
|
|
1D6P
 
 | | HUMAN LYSOZYME L63 MUTANT LABELLED WITH 2',3'-EPOXYPROPYL N,N'-DIACETYLCHITOBIOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LYSOZYME | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1999-10-15 | | Release date: | 2000-01-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Protein-carbohydrate interactions in human lysozyme probed by combining site-directed mutagenesis and affinity labeling.
Biochemistry, 39, 2000
|
|
5AWL
 
 | | CRYSTAL STRUCTURE OF A MUTANT OF CHIGNOLIN, CLN025 | | Descriptor: | A mutant of Chignolin, CLN025 | | Authors: | Akiba, T, Ishimura, M, Odahara, T, Harata, K, Honda, S. | | Deposit date: | 2015-07-05 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Crystal structure of a ten-amino acid protein
J.Am.Chem.Soc., 130, 2008
|
|
2O4A
 
 | |
2O49
 
 | |
1I75
 
 | | CRYSTAL STRUCTURE OF CYCLODEXTRIN GLUCANOTRANSFERASE FROM ALKALOPHILIC BACILLUS SP.#1011 COMPLEXED WITH 1-DEOXYNOJIRIMYCIN | | Descriptor: | 1-DEOXYNOJIRIMYCIN, CALCIUM ION, CYCLODEXTRIN GLUCANOTRANSFERASE | | Authors: | Kanai, R, Haga, K, Yamane, K, Harata, K. | | Deposit date: | 2001-03-08 | | Release date: | 2001-04-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011 complexed with 1-deoxynojirimycin at 2.0 A resolution.
J.Biochem.(Tokyo), 129, 2001
|
|
1K7U
 
 | | Crystal Structure Analysis of crosslinked-WGA3/GlcNAcbeta1,4GlcNAc complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, agglutinin isolectin 3 | | Authors: | Muraki, M, Ishimura, M, Harata, K. | | Deposit date: | 2001-10-22 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions of wheat-germ agglutinin with GlcNAc beta 1,6Gal sequence
Biochim.Biophys.Acta, 1569, 2002
|
|
1K7V
 
 | | Crystal Structure Analysis of crosslinked-WGA3/GlcNAcbeta1,6Galbeta1,4Glc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, agglutinin isolectin 3 | | Authors: | Muraki, M, Ishimura, M, Harata, K. | | Deposit date: | 2001-10-22 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions of wheat-germ agglutinin with GlcNAc beta 1,6Gal sequence
Biochim.Biophys.Acta, 1569, 2002
|
|
1K7T
 
 | | Crystal Structure Analysis of crosslinked-WGA3/GlcNAcbeta1,6Gal complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-beta-D-galactopyranose, agglutinin isolectin 3 | | Authors: | Muraki, M, Ishimura, M, Harata, K. | | Deposit date: | 2001-10-22 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interactions of wheat-germ agglutinin with GlcNAc beta 1,6Gal sequence
Biochim.Biophys.Acta, 1569, 2002
|
|
1GD9
 
 | | CRYSTALL STRUCTURE OF PYROCOCCUS PROTEIN-A1 | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Harata, K, Matsui, I, Kuramitsu, S. | | Deposit date: | 2000-09-22 | | Release date: | 2001-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature dependence of the enzyme-substrate recognition mechanism.
J.Biochem., 129, 2001
|
|
1GDE
 
 | | CRYSTAL STRUCTURE OF PYROCOCCUS PROTEIN A-1 E-FORM | | Descriptor: | ASPARTATE AMINOTRANSFERASE, GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Harata, K, Matsui, I, Kuramitsu, S. | | Deposit date: | 2000-09-23 | | Release date: | 2001-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature dependence of the enzyme-substrate recognition mechanism.
J.Biochem., 129, 2001
|
|
1D7F
 
 | | CRYSTAL STRUCTURE OF ASPARAGINE 233-REPLACED CYCLODEXTRIN GLUCANOTRANSFERASE FROM ALKALOPHILIC BACILLUS SP. 1011 DETERMINED AT 1.9 A RESOLUTION | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLUCANOTRANSFERASE | | Authors: | Ishii, N, Haga, K, Yamane, K, Harata, K. | | Deposit date: | 1999-10-18 | | Release date: | 2000-03-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of asparagine 233-replaced cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011 determined at 1.9 A resolution.
J.Mol.Recog., 13, 2000
|
|
1DED
 
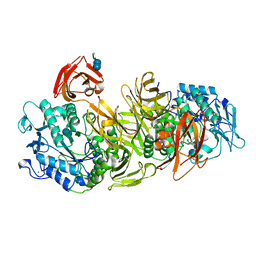 | | CRYSTAL STRUCTURE OF ALKALOPHILIC ASPARAGINE 233-REPLACED CYCLODEXTRIN GLUCANOTRANSFERASE COMPLEXED WITH AN INHIBITOR, ACARBOSE, AT 2.0 A RESOLUTION | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, CYCLODEXTRIN GLUCANOTRANSFERASE | | Authors: | Ishii, N, Haga, K, Yamane, K, Harata, K. | | Deposit date: | 1999-11-14 | | Release date: | 2000-04-07 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alkalophilic asparagine 233-replaced cyclodextrin glucanotransferase complexed with an inhibitor, acarbose, at 2.0 A resolution.
J.Biochem.(Tokyo), 127, 2000
|
|
2TSB
 
 | | AZURIN MUTANT M121A-AZIDE | | Descriptor: | AZIDE ION, AZURIN AZIDE, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2TSA
 
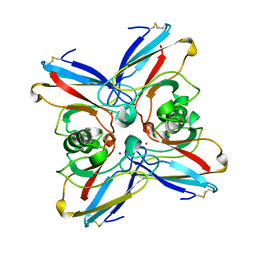 | | AZURIN MUTANT M121A | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1WP6
 
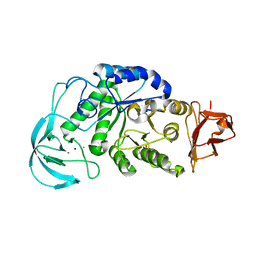 | | Crystal structure of maltohexaose-producing amylase from alkalophilic Bacillus sp.707. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Glucan 1,4-alpha-maltohexaosidase, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2004-08-31 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and crystallographic analyses of maltohexaose-producing amylase from alkalophilic Bacillus sp. 707
Biochemistry, 43, 2004
|
|
1WPC
 
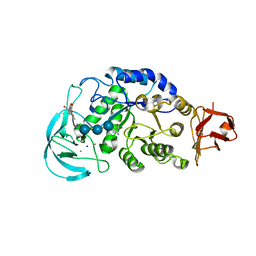 | | Crystal structure of maltohexaose-producing amylase complexed with pseudo-maltononaose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2004-09-01 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and crystallographic analyses of maltohexaose-producing amylase from alkalophilic Bacillus sp. 707
Biochemistry, 43, 2004
|
|
2D3L
 
 | | Crystal structure of maltohexaose-producing amylase from Bacillus sp.707 complexed with maltopentaose. | | Descriptor: | CALCIUM ION, Glucan 1,4-alpha-maltohexaosidase, SODIUM ION, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2005-09-29 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Trp140 at subsite -6 on the maltohexaose production of maltohexaose-producing amylase from alkalophilic Bacillus sp.707
Protein Sci., 15, 2006
|
|
2D3N
 
 | | Crystal structure of maltohexaose-producing amylase from Bacillus sp.707 complexed with maltohexaose | | Descriptor: | CALCIUM ION, Glucan 1,4-alpha-maltohexaosidase, SODIUM ION, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2005-09-29 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Trp140 at subsite -6 on the maltohexaose production of maltohexaose-producing amylase from alkalophilic Bacillus sp.707
Protein Sci., 15, 2006
|
|
1V3M
 
 | | Crystal structure of F283Y mutant cyclodextrin glycosyltransferase complexed with a pseudo-tetraose derived from acarbose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2003-11-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Phe283 in enzymatic reaction of cyclodextrin glycosyltransferase from alkalophilic Bacillus sp.1011: Substrate binding and arrangement of the catalytic site
PROTEIN SCI., 13, 2004
|
|
