3OE9
 
 | | Crystal structure of the chemokine CXCR4 receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
1VPZ
 
 | |
1VQR
 
 | |
1VR8
 
 | |
1VK3
 
 | |
1VL4
 
 | |
1VLP
 
 | |
1VKH
 
 | |
3ODU
 
 | | The 2.5 A structure of the CXCR4 chemokine receptor in complex with small molecule antagonist IT1t | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OE6
 
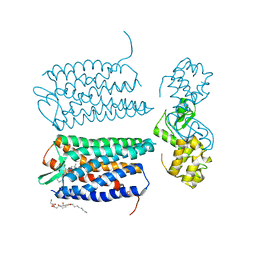 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in I222 spacegroup | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OE8
 
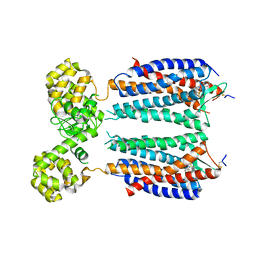 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
1ZCZ
 
 | |
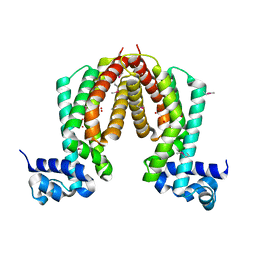
1ZKG
 
 | |
1ZX8
 
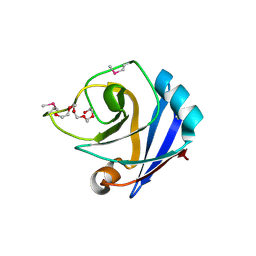 | |
3OE0
 
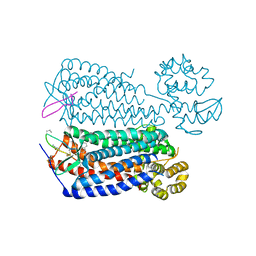 | | Crystal structure of the CXCR4 chemokine receptor in complex with a cyclic peptide antagonist CVX15 | | Descriptor: | C-X-C chemokine receptor type 4, Lysozyme Chimera, Polyphemusin analog, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3H41
 
 | |
3H50
 
 | |
3HSA
 
 | |
3IRB
 
 | |
3H0N
 
 | |
3HBZ
 
 | |
3K5J
 
 | |
3BOS
 
 | |
3CM1
 
 | |
3DEE
 
 | |
