7NXL
 
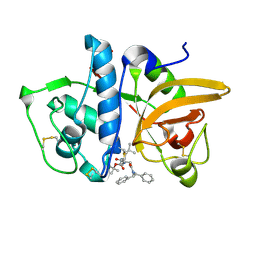 | | Structure of human cathepsin K in complex with the acrylamide inhibitor Gu3110 | | Descriptor: | Cathepsin K, SULFATE ION, tert-butyl (1-((4-(dibenzylamino)-4-oxobutyl)amino)-4-methyl-1-oxopentan-2-yl)carbamate | | Authors: | Busa, M, Benysek, J, Lemke, C, Gutschow, M, Mares, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-09-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Activity-Based Probe for Cathepsin K Imaging with Excellent Potency and Selectivity.
J.Med.Chem., 64, 2021
|
|
7NXM
 
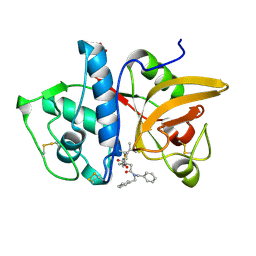 | | Structure of human cathepsin K in complex with the selective activity-based probe Gu3416 | | Descriptor: | Cathepsin K, N-(4-(dibenzylamino)-4-oxobutyl)-2-(5-(dimethylamino)pentanamido)-4-methylpentanamide, SULFATE ION | | Authors: | Busa, M, Benysek, J, Lemke, C, Gutschow, M, Mares, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | An Activity-Based Probe for Cathepsin K Imaging with Excellent Potency and Selectivity.
J.Med.Chem., 64, 2021
|
|
8CQL
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-N-((S)-1-(5-Fluoro-2-methoxy-4-(4-methylthiazol-5-yl)phenyl)ethyl)-1-((S)-2-(1-fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxamide (Compound 33) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-[5-fluoranyl-2-methoxy-4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Casement, R, Phuong Vu, L, Ciulli, A, Gutschow, M. | | Deposit date: | 2023-03-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Expanding the Structural Diversity at the Phenylene Core of Ligands for the von Hippel-Lindau E3 Ubiquitin Ligase: Development of Highly Potent Hypoxia-Inducible Factor-1 alpha Stabilizers.
J.Med.Chem., 66, 2023
|
|
8CQK
 
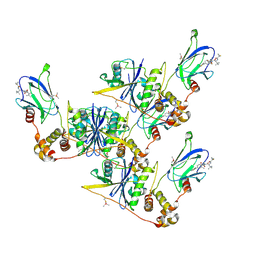 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(1-Fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(2-methyl-4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (Compound 30) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-[2-methyl-4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Casement, R, Phuong Vu, L, Ciulli, A, Gutschow, M. | | Deposit date: | 2023-03-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Expanding the Structural Diversity at the Phenylene Core of Ligands for the von Hippel-Lindau E3 Ubiquitin Ligase: Development of Highly Potent Hypoxia-Inducible Factor-1 alpha Stabilizers.
J.Med.Chem., 66, 2023
|
|
8CQE
 
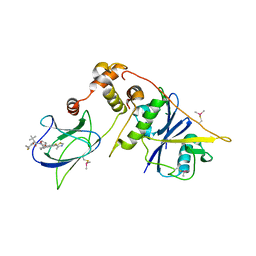 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(1-Fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(2-methyl-4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (Compound 37) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-7-fluoranyl-6-(4-methyl-1,3-thiazol-5-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Casement, R, Phuong Vu, L, Ciulli, A, Gutschow, M. | | Deposit date: | 2023-03-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Expanding the Structural Diversity at the Phenylene Core of Ligands for the von Hippel-Lindau E3 Ubiquitin Ligase: Development of Highly Potent Hypoxia-Inducible Factor-1 alpha Stabilizers.
J.Med.Chem., 66, 2023
|
|
6YI7
 
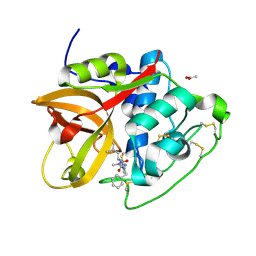 | | Structure of cathepsin B1 from Schistosoma mansoni (SmCB1) in complex with an azanitrile inhibitor | | Descriptor: | 1-[(2~{S})-1-[[iminomethyl(methyl)amino]-methyl-amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-3-(phenylmethyl)urea, ACETATE ION, Cathepsin B-like peptidase (C01 family) | | Authors: | Jilkova, A, Rezacova, P, Pachl, P, Fanfrlik, J, Rubesova, P, Guetschow, M, Mares, M. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Azanitrile Inhibitors of the SmCB1 Protease Target Are Lethal to Schistosoma mansoni : Structural and Mechanistic Insights into Chemotype Reactivity.
Acs Infect Dis., 7, 2021
|
|
6F5M
 
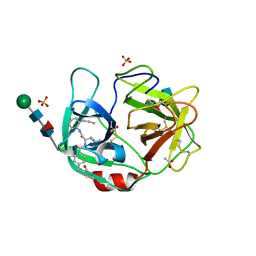 | | Crystal structure of highly glycosylated human leukocyte elastase in complex with a thiazolidinedione inhibitor | | Descriptor: | 5-[[4-[[(2~{S})-4-methyl-1-oxidanylidene-1-[(2-propylphenyl)amino]pentan-2-yl]carbamoyl]phenyl]methyl]-2-oxidanylidene-1,3-thiazol-1-ium-4-olate, ACETATE ION, Neutrophil elastase, ... | | Authors: | Hochscherf, J, Pietsch, M, Tieu, W, Kuan, K, Hautmann, S, Abell, A, Guetschow, M, Niefind, K. | | Deposit date: | 2017-12-01 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of highly glycosylated human leukocyte elastase in complex with an S2' site binding inhibitor.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7E18
 
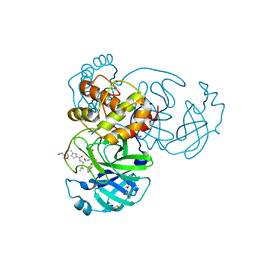 | | Crystal structure of SAR-CoV-2 3CL protease complex with inhibitor YH-53 | | Descriptor: | 1,2-ETHANEDIOL, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Replicase polyprotein 1ab | | Authors: | Senda, M, Konno, S, Hayashi, Y, Senda, T. | | Deposit date: | 2021-02-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 3CL Protease Inhibitors with an Electrophilic Arylketone Moiety as Anti-SARS-CoV-2 Agents.
J.Med.Chem., 65, 2022
|
|
7E19
 
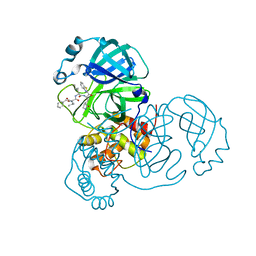 | | Crystal structure of SAR-CoV-2 3CL protease complex with inhibitor SH-5 | | Descriptor: | (phenylmethyl) N-[(2S)-1-[[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]carbamate, 3C-like proteinase | | Authors: | Senda, M, Konno, S, Hayashi, Y, Senda, T. | | Deposit date: | 2021-02-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3CL Protease Inhibitors with an Electrophilic Arylketone Moiety as Anti-SARS-CoV-2 Agents.
J.Med.Chem., 65, 2022
|
|
4Q2K
 
 | | Bovine alpha chymotrypsin bound to a cyclic peptide inhibitor, 5b | | Descriptor: | (11S)-4,9-dioxo-N-[(2S)-1-oxo-3-phenylpropan-2-yl]-17,22-dioxa-10,30-diazatetracyclo[21.2.2.2~13,16~.1~5,8~]triaconta-1(25),5,7,13,15,23,26,28-octaene-11-carboxamide, Chymotrypsinogen A | | Authors: | Chan, H.Y, Bruning, J.B, Abell, A.D. | | Deposit date: | 2014-04-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Macrocyclic protease inhibitors with reduced peptide character.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8ACD
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GA-17S | | Descriptor: | (2~{S})-4-[[2,4-bis(oxidanylidene)-1~{H}-pyrimidin-6-yl]carbonyl]-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
8ACL
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GC-14 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-pyridin-3-ylcarbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
7QBN
 
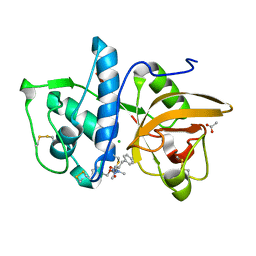 | | Structure of cathepsin K in complex with the azadipeptide nitrile inhibitor Gu1303 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[aminomethyl(methyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7QBM
 
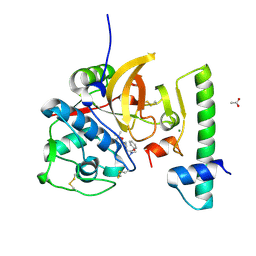 | | Structure of the activation intermediate of cathepsin K in complex with the 3-cyano-3-aza-beta-amino acid inhibitor Gu2602 | | Descriptor: | ACETATE ION, Cathepsin K, MAGNESIUM ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7QBL
 
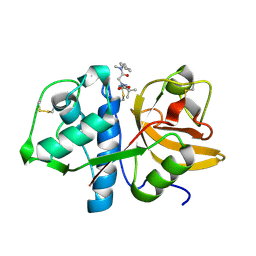 | |
7QBO
 
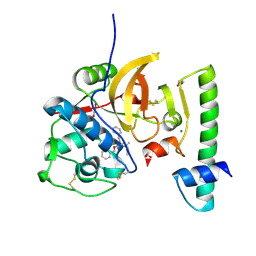 | | Structure of the activation intermediate of cathepsin K in complex with the azadipeptide nitrile inhibitor Gu1303 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[aminomethyl(methyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
8B56
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the inhibitor GD-9 | | Descriptor: | (2~{S})-4-(2-chloranylethanoyl)-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5, BROMIDE ION, ... | | Authors: | Straeter, N, Muller, C.E, Claff, T, Sylvester, K, Weisse, R, Gao, S, Song, L, Liu, X, Zhan, P. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Discovery and Crystallographic Studies of Nonpeptidic Piperazine Derivatives as Covalent SARS-CoV-2 Main Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7X6K
 
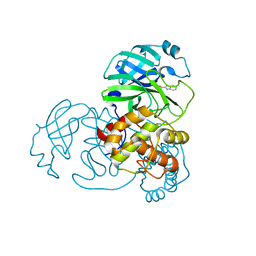 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3w | | Descriptor: | 1H-indole-2-carbaldehyde, 3C-like proteinase | | Authors: | Su, H, Nie, T, Xie, H, Li, Z.W, Li, M.J, Xu, Y. | | Deposit date: | 2022-03-07 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Small-Molecule Thioesters as SARS-CoV-2 Main Protease Inhibitors: Enzyme Inhibition, Structure-Activity Relationships, Antiviral Activity, and X-ray Structure Determination.
J.Med.Chem., 65, 2022
|
|
7X6J
 
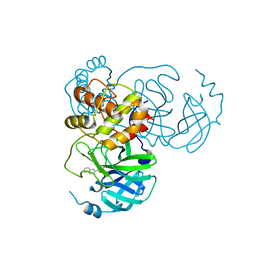 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3af | | Descriptor: | 3C-like proteinase, quinoline-2-carboxylic acid | | Authors: | Su, H, Nie, T, Xie, H, Li, Z.W, Li, M.J, Xu, Y. | | Deposit date: | 2022-03-07 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small-Molecule Thioesters as SARS-CoV-2 Main Protease Inhibitors: Enzyme Inhibition, Structure-Activity Relationships, Antiviral Activity, and X-ray Structure Determination.
J.Med.Chem., 65, 2022
|
|
8OU3
 
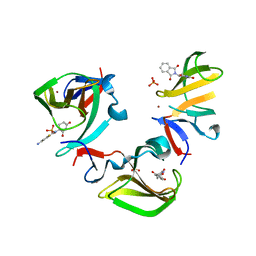 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 8d | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-fluoranyl-benzamide, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Heim, C, Bischof, L, Maiwald, S, Hartmann, M.D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU5
 
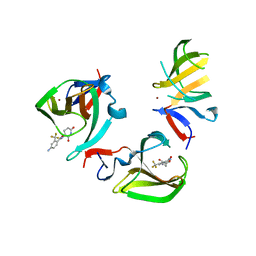 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11b | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-(trifluoromethyl)benzamide, Cereblon isoform 4, ZINC ION | | Authors: | Heim, C, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU4
 
 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11a | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-chloranyl-benzamide, Cereblon isoform 4, ZINC ION | | Authors: | Heim, C, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU7
 
 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11d | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-methoxy-benzamide, Cereblon isoform 4, ZINC ION | | Authors: | Heim, C, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-04-22 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU9
 
 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11e | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-oxidanyl-benzamide, Cereblon isoform 4, ZINC ION | | Authors: | Heim, C, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-04-22 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU6
 
 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11c | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-methyl-benzamide, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Heim, C, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
