4ZXH
 
 | | Crystal Structure of holo-AB3403 a four domain nonribosomal peptide synthetase from Acinetobacter Baumanii | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4'-PHOSPHOPANTETHEINE, ABBFA_003403, ... | | Authors: | Drake, E.J, Miller, B.R, Allen, C.L, Gulick, A.M. | | Deposit date: | 2015-05-20 | | Release date: | 2015-12-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of two distinct conformations of holo-non-ribosomal peptide synthetases.
Nature, 529, 2016
|
|
8G98
 
 | |
8G97
 
 | |
8G95
 
 | |
8G96
 
 | |
2FQ1
 
 | | Crystal structure of the two-domain non-ribosomal peptide synthetase EntB containing isochorismate lyase and aryl-carrier protein domains | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Isochorismatase, ... | | Authors: | Drake, E.J, Nicolai, D.A, Gulick, A.M. | | Deposit date: | 2006-01-17 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the EntB multidomain nonribosomal peptide synthetase and functional analysis of its interaction with the EntE adenylation domain.
Chem.Biol., 13, 2006
|
|
7TGK
 
 | | Crystal structure of ATP bound DesD, the desferrioxamine synthetase from the Streptomyces griseoflavus ferrimycin biosynthetic pathway | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Patel, K.D, Gulick, A.M. | | Deposit date: | 2022-01-07 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An acyl-adenylate mimic reveals the structural basis for substrate recognition by the iterative siderophore synthetase DesD.
J.Biol.Chem., 298, 2022
|
|
7TGJ
 
 | |
7TGL
 
 | |
7TGN
 
 | | Crystal structure of DesD, the desferrioxamine synthetase from the Streptomyces violaceus salmycin biosynthetic pathway | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Desferrioxamine synthetase DesD, ... | | Authors: | Patel, K.D, Gulick, A.M. | | Deposit date: | 2022-01-07 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An acyl-adenylate mimic reveals the structural basis for substrate recognition by the iterative siderophore synthetase DesD.
J.Biol.Chem., 298, 2022
|
|
7TGM
 
 | |
4OOF
 
 | | M. tuberculosis 1-deoxy-d-xylulose-5-phosphate reductoisomerase W203F mutant bound to fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Allen, C.L, Kholodar, S.A, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alteration of the Flexible Loop in 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase Boosts Enthalpy-Driven Inhibition by Fosmidomycin.
Biochemistry, 53, 2014
|
|
4OOE
 
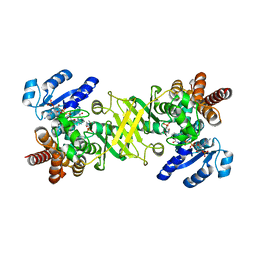 | | M. tuberculosis 1-deoxy-d-xylulose-5-phosphate reductoisomerase W203Y mutant bound to fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Allen, C.L, Kholodar, S.A, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.826 Å) | | Cite: | Alteration of the Flexible Loop in 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase Boosts Enthalpy-Driven Inhibition by Fosmidomycin.
Biochemistry, 53, 2014
|
|
6C4C
 
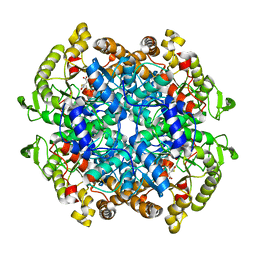 | | Crystal structure of 3-nitropropionate modified isocitrate lyase from Mycobacterium tuberculosis with glyoxylate and pyruvate | | Descriptor: | 3-NITROPROPANOIC ACID, GLYOXYLIC ACID, Isocitrate lyase 1, ... | | Authors: | Kreitler, D.F, Ray, S, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Nitro Group as a Masked Electrophile in Covalent Enzyme Inhibition.
ACS Chem. Biol., 13, 2018
|
|
6CN7
 
 | |
6E8Z
 
 | |
6E8Y
 
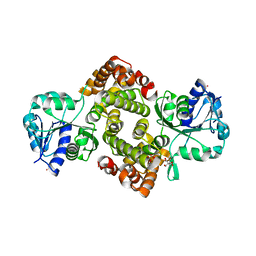 | | Unliganded Human Glycerol 3-Phosphate Dehydrogenase | | Descriptor: | 2,2-bis(hydroxymethyl)propane-1,3-diol, Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic, ... | | Authors: | Mydy, L.S, Gulick, A.M. | | Deposit date: | 2018-07-31 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Human Glycerol 3-Phosphate Dehydrogenase: X-ray Crystal Structures That Guide the Interpretation of Mutagenesis Studies.
Biochemistry, 58, 2019
|
|
6E90
 
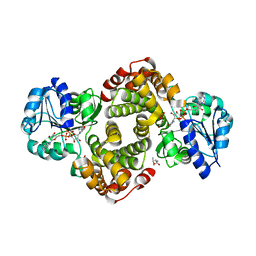 | | Ternary complex of human glycerol 3-phosphate dehydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, CALCIUM ION, ... | | Authors: | Mydy, L.S, Gulick, A.M. | | Deposit date: | 2018-07-31 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Human Glycerol 3-Phosphate Dehydrogenase: X-ray Crystal Structures That Guide the Interpretation of Mutagenesis Studies.
Biochemistry, 58, 2019
|
|
5JA1
 
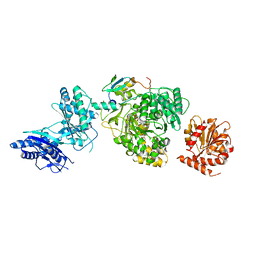 | | EntF, a Terminal Nonribosomal Peptide Synthetase Module Bound to the MbtH-Like Protein YbdZ | | Descriptor: | 5'-({[(2R,3S)-3-amino-4-hydroxy-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}butyl]sulfonyl}amino)-5'-deoxyadenosine, CHLORIDE ION, Enterobactin biosynthesis protein YbdZ, ... | | Authors: | Miller, B.R, Gulick, A.M. | | Deposit date: | 2016-04-11 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of a Nonribosomal Peptide Synthetase Module Bound to MbtH-like Proteins Support a Highly Dynamic Domain Architecture.
J.Biol.Chem., 291, 2016
|
|
5JA2
 
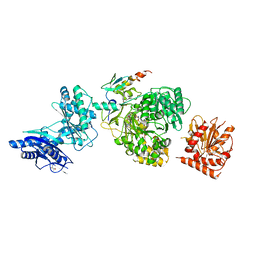 | | EntF, a Terminal Nonribosomal Peptide Synthetase Module Bound to the non-Native MbtH-Like Protein PA2412 | | Descriptor: | 5'-({[(2R,3S)-3-amino-4-hydroxy-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}butyl]sulfonyl}amino)-5'-deoxyadenosine, Enterobactin synthase component F, MbtH-Like Protein PA2412, ... | | Authors: | Miller, B.R, Gulick, A.M. | | Deposit date: | 2016-04-11 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of a Nonribosomal Peptide Synthetase Module Bound to MbtH-like Proteins Support a Highly Dynamic Domain Architecture.
J.Biol.Chem., 291, 2016
|
|
3CW8
 
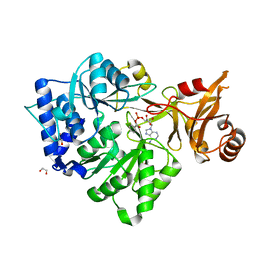 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, bound to 4CBA-Adenylate | | Descriptor: | 1,2-ETHANEDIOL, 4-chlorobenzoyl CoA ligase, 5'-O-[(S)-{[(4-chlorophenyl)carbonyl]oxy}(hydroxy)phosphoryl]adenosine | | Authors: | Reger, A.S, Cao, J, Wu, R, Dunaway-Mariano, D, Gulick, A.M. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
Biochemistry, 47, 2008
|
|
3L94
 
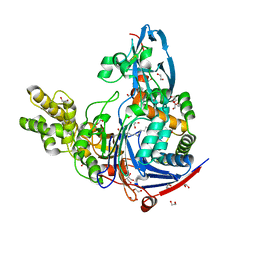 | | Structure of PvdQ covalently acylated with myristate | | Descriptor: | 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase pvdQ subunit alpha, Acyl-homoserine lactone acylase pvdQ subunit beta, ... | | Authors: | Drake, E.J, Gulick, A.M. | | Deposit date: | 2010-01-04 | | Release date: | 2011-01-26 | | Last modified: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization and high-throughput screening of inhibitors of PvdQ, an NTN hydrolase involved in pyoverdine synthesis.
Acs Chem.Biol., 6, 2011
|
|
3L91
 
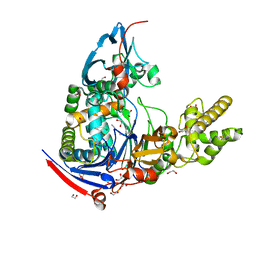 | | Structure of Pseudomonas aerugionsa PvdQ bound to octanoate | | Descriptor: | 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase pvdQ subunit alpha, Acyl-homoserine lactone acylase pvdQ subunit beta, ... | | Authors: | Drake, E.J, Gulick, A.M. | | Deposit date: | 2010-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural characterization and high-throughput screening of inhibitors of PvdQ, an NTN hydrolase involved in pyoverdine synthesis.
Acs Chem.Biol., 6, 2011
|
|
3O83
 
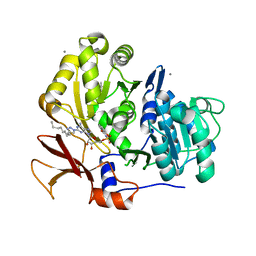 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 2-(4-n-dodecyl-1,2,3-triazol-1-yl)-5'-O-[N-(2-hydroxybenzoyl)sulfamoyl]adenosine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-dodecyl-1H-1,2,3-triazol-1-yl)-5'-O-{[(2-hydroxyphenyl)carbonyl]sulfamoyl}adenosine, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
4DG8
 
 | | Structure of PA1221, an NRPS protein containing adenylation and PCP domains | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE MONOPHOSPHATE, PA1221 | | Authors: | Mitchell, C.A, Shi, C, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2012-01-25 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of PA1221, a Nonribosomal Peptide Synthetase Containing Adenylation and Peptidyl Carrier Protein Domains.
Biochemistry, 51, 2012
|
|
