2LVL
 
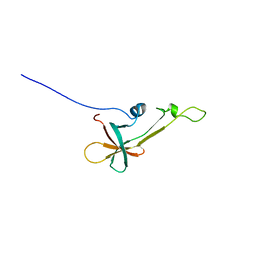 | | NMR Structure the lantibiotic immunity protein SpaI | | Descriptor: | SpaI | | Authors: | Christ, N, Bochmann, S, Gottstein, D, Duchardt-Ferner, E, Hellmich, U.A, Duesterhus, S, Koetter, P, Guentert, P, Entian, K, Woehnert, J. | | Deposit date: | 2012-07-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The First Structure of a Lantibiotic Immunity Protein, SpaI from Bacillus subtilis, Reveals a Novel Fold.
J.Biol.Chem., 287, 2012
|
|
2MD9
 
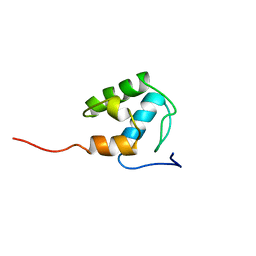 | | Solution Structure of an Active Site Mutant Pepitdyl Carrier Protein | | Descriptor: | Tyrocidine synthase 3 | | Authors: | Tufar, P, Rahighi, S, Kraas, F.I, Kirchner, D.K, Loehr, F, Henrich, E, Koepke, J, Dikic, I, Guentert, P, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Crystal Structure of a PCP/Sfp Complex Reveals the Structural Basis for Carrier Protein Posttranslational Modification.
Chem.Biol., 21, 2014
|
|
2LUE
 
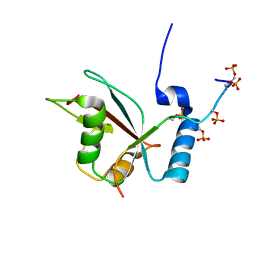 | | LC3B OPTN-LIR Ptot complex structure | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Optineurin | | Authors: | Rogov, V.V, Rozenknop, A, Loehr, F, Guentert, P, Doetsch, V. | | Deposit date: | 2012-06-13 | | Release date: | 2013-07-17 | | Last modified: | 2022-08-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella.
Biochem.J., 454, 2013
|
|
2DT7
 
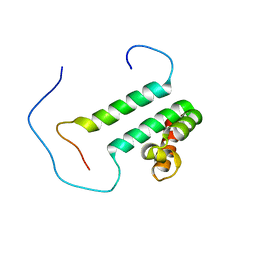 | | Solution structure of the second SURP domain of human splicing factor SF3a120 in complex with a fragment of human splicing factor SF3a60 | | Descriptor: | Splicing factor 3 subunit 1, Splicing factor 3A subunit 3 | | Authors: | He, F, Kuwasako, K, Inoue, M, Guntert, P, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-11 | | Release date: | 2006-12-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SURP domains and the subunit-assembly mechanism within the splicing factor SF3a complex in 17S U2 snRNP
Structure, 14, 2006
|
|
2L8J
 
 | | GABARAPL-1 NBR1-LIR complex structure | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, NBR1-LIR peptide | | Authors: | Rogov, V.V, Rozenknop, A, Rogova, N.Y, Loehr, F, Guentert, P, Dikic, I, Doetsch, V. | | Deposit date: | 2011-01-17 | | Release date: | 2011-05-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Interaction of GABARAPL-1 with the LIR Motif of NBR1.
J.Mol.Biol., 410, 2011
|
|
2MN9
 
 | |
2MN8
 
 | |
2MMJ
 
 | |
2NB1
 
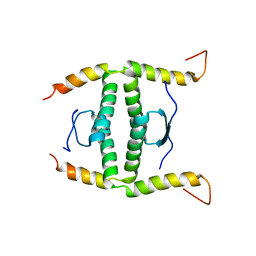 | | P63/p73 hetero-tetramerisation domain | | Descriptor: | Tumor protein 63, Tumor protein p73 | | Authors: | Gebel, J, Buchner, L, Loehr, F.M, Luh, L.M, Coutandin, D, Guentert, P, Doetsch, V. | | Deposit date: | 2016-01-19 | | Release date: | 2016-12-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Mechanism of TAp73 inhibition by Delta Np63 and structural basis of p63/p73 hetero-tetramerization.
Cell Death Differ., 23, 2016
|
|
2MVX
 
 | | Atomic-resolution 3D structure of amyloid-beta fibrils: the Osaka mutation | | Descriptor: | Amyloid beta A4 protein | | Authors: | Schuetz, A.K, Vagt, T, Huber, M, Ovchinnikova, O.Y, Cadalbert, R, Wall, J, Guentert, P, Bockmann, A, Glockshuber, R, Meier, B.H. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-26 | | Last modified: | 2015-01-21 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of Amyloid beta Fibrils Bearing the Osaka Mutation.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2N5E
 
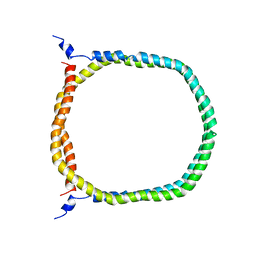 | | The 3D solution structure of discoidal high-density lipoprotein particles | | Descriptor: | Apolipoprotein A-I | | Authors: | Bibow, S, Polyhach, Y, Eichmann, C, Chi, C.N, Kowal, J, Stahlberg, H, Jeschke, G, Guentert, P, Riek, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-12-21 | | Last modified: | 2017-02-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I.
Nat.Struct.Mol.Biol., 24, 2017
|
|
2MYX
 
 | | Structure of the CUE domain of yeast Cue1 | | Descriptor: | Coupling of ubiquitin conjugation to ER degradation protein 1 | | Authors: | Kniss, A, Rogov, V.V, Loehr, F, Guentert, P, Doetsch, V. | | Deposit date: | 2015-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The CUE Domain of Cue1 Aligns Growing Ubiquitin Chains with Ubc7 for Rapid Elongation.
Mol.Cell, 62, 2016
|
|
2ROG
 
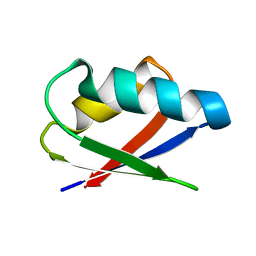 | | Solution structure of Thermus thermophilus HB8 TTHA1718 protein in living E. coli cells | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2009-03-03 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
2RON
 
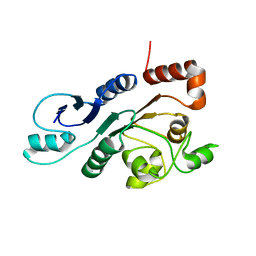 | | The external thioesterase of the Surfactin-Synthetase | | Descriptor: | Surfactin synthetase thioesterase subunit | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guentert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-12 | | Last modified: | 2016-10-26 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase
Nature, 454, 2008
|
|
2ROE
 
 | | Solution structure of thermus thermophilus HB8 TTHA1718 protein in vitro | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-20 | | Release date: | 2009-03-03 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
1G6X
 
 | | ULTRA HIGH RESOLUTION STRUCTURE OF BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) MUTANT WITH ALTERED BINDING LOOP SEQUENCE | | Descriptor: | 1,2-ETHANEDIOL, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Addlagatta, A, Czapinska, H, Krzywda, S, Otlewski, J, Jaskolski, M. | | Deposit date: | 2000-11-08 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Ultrahigh-resolution structure of a BPTI mutant.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1QLQ
 
 | | Bovine Pancreatic Trypsin Inhibitor (BPTI) Mutant with Altered Binding Loop Sequence | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Czapinska, H, Krzywda, S, Sheldrick, G.M, Otlewski, J, Jaskolski, M. | | Deposit date: | 1999-09-10 | | Release date: | 1999-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High Resolution Structure of Bovine Pancreatic Trypsin Inhibitor with Altered Binding Loop Sequence
J.Mol.Biol., 295, 1999
|
|
