4MT9
 
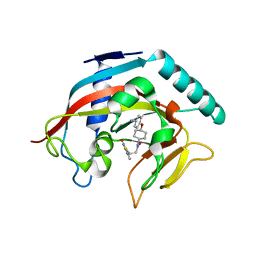 | | Co-crystal structure of tankyrase 1 with compound 49 | | 分子名称: | N-[trans-4-(4-cyanophenoxy)cyclohexyl]-3-[(4-oxo-3,4-dihydroquinazolin-2-yl)sulfanyl]propanamide, Tankyrase-1, ZINC ION | | 著者 | Huang, X. | | 登録日 | 2013-09-19 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4MSG
 
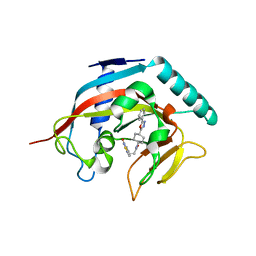 | | Crystal structure of tankyrase 1 with compound 22 | | 分子名称: | 3-[(4-oxo-3,4-dihydroquinazolin-2-yl)sulfanyl]-N-[trans-4-(5-phenyl-1,3,4-oxadiazol-2-yl)cyclohexyl]propanamide, Tankyrase-1, ZINC ION | | 著者 | Huang, X. | | 登録日 | 2013-09-18 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
8DEY
 
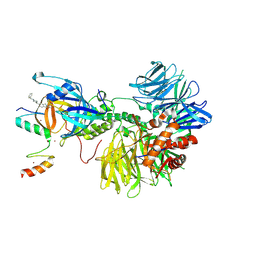 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2,3) and the molecular glue DKY709 | | 分子名称: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Ma, X, Ornelas, E, Clifton, M.C. | | 登録日 | 2022-06-21 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
7T5U
 
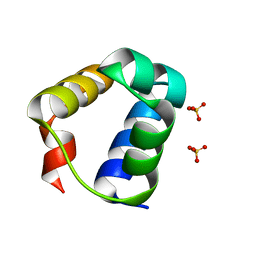 | |
7T5T
 
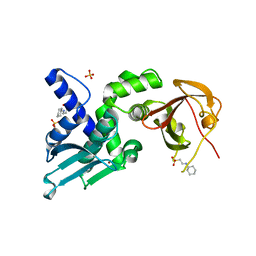 | | Structure of Thauera sp. K11 CapP | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CapP toxin, SULFATE ION, ... | | 著者 | Lau, R.K, Corbett, K.D. | | 登録日 | 2021-12-13 | | 公開日 | 2022-09-14 | | 最終更新日 | 2022-11-30 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | A conserved signaling pathway activates bacterial CBASS immune signaling in response to DNA damage.
Embo J., 41, 2022
|
|
7T5W
 
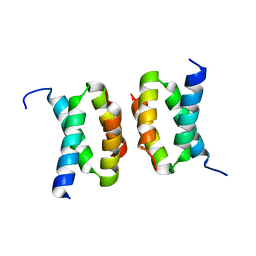 | |
7T5V
 
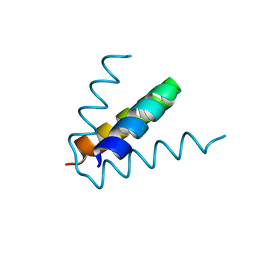 | |
7U8F
 
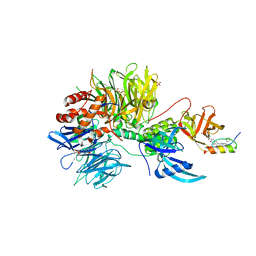 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2) and the molecular glue DKY709 | | 分子名称: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA damage-binding protein 1, ... | | 著者 | Ma, X, Ornelas, E, Clifton, M.C. | | 登録日 | 2022-03-08 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
7UBA
 
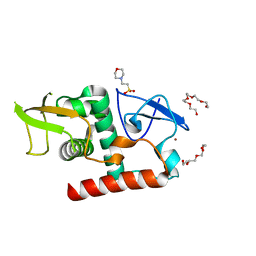 | | Structure of fungal Hop1 CBR domain | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HORMA domain-containing protein, PENTAETHYLENE GLYCOL, ... | | 著者 | Ur, S.N, Corbett, K.D. | | 登録日 | 2022-03-14 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Chromatin binding by HORMAD proteins regulates meiotic recombination initiation.
Embo J., 43, 2024
|
|
6LHC
 
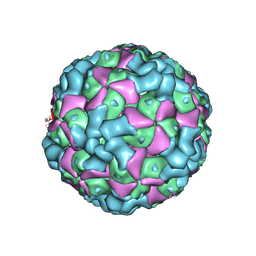 | | The cryo-EM structure of coxsackievirus A16 empty particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-07 | | 公開日 | 2020-02-05 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.43 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHL
 
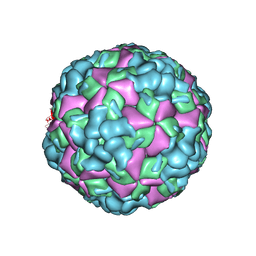 | | The cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab 18A7 | | 分子名称: | VP1 protein, VP2 protein, VP3 protein | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-09 | | 公開日 | 2020-02-05 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHB
 
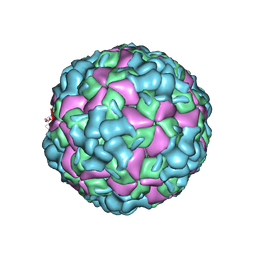 | | The cryo-EM structure of coxsackievirus A16 A-particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-07 | | 公開日 | 2020-02-05 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHP
 
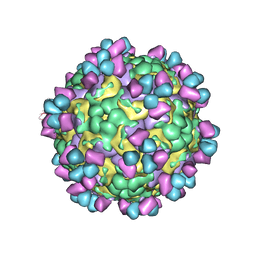 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 14B10 | | 分子名称: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-09 | | 公開日 | 2020-02-05 | | 最終更新日 | 2020-02-26 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHO
 
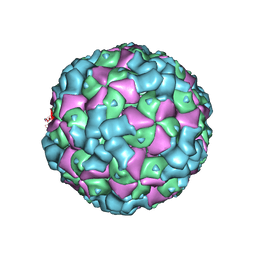 | | The cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab 18A7 | | 分子名称: | VP1 protein, VP2 protein, VP3 protein | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-09 | | 公開日 | 2020-02-05 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHK
 
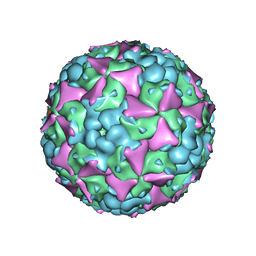 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 18A7 | | 分子名称: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-09 | | 公開日 | 2020-02-05 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (2.65 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHA
 
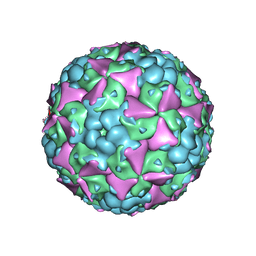 | | The cryo-EM structure of coxsackievirus A16 mature virion | | 分子名称: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-07 | | 公開日 | 2020-02-05 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.56 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHQ
 
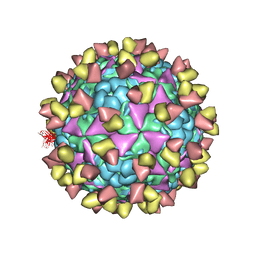 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab NA9D7 | | 分子名称: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-09 | | 公開日 | 2020-02-05 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHT
 
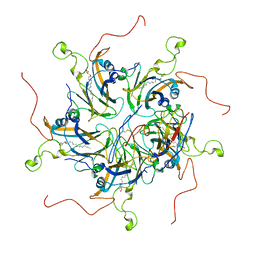 | | Localized reconstruction of coxsackievirus A16 mature virion in complex with Fab 18A7 | | 分子名称: | SPHINGOSINE, VP1 protein, heavy chain variable region of Fab 18A7, ... | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-10 | | 公開日 | 2020-02-05 | | 最終更新日 | 2020-02-26 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
2OSC
 
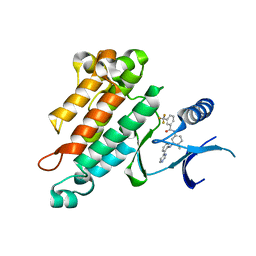 | | Synthesis, Structural Analysis, and SAR Studies of Triazine Derivatives as Potent, Selective Tie-2 Inhibitors | | 分子名称: | Angiopoietin-1 receptor, N-{4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]PHENYL}-3-(TRIFLUOROMETHYL)BENZAMIDE | | 著者 | Bellon, S.F, Kim, J. | | 登録日 | 2007-02-05 | | 公開日 | 2007-03-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Synthesis, structural analysis, and SAR studies of triazine derivatives as potent, selective Tie-2 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPF
 
 | |
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | 登録日 | 2020-12-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | 分子名称: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
