3TJD
 
 | | co-crystal structure of Jak2 with thienopyridine 19 | | 分子名称: | 4-amino-2-[4-(tert-butylsulfamoyl)phenyl]-N-methylthieno[3,2-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | 著者 | Huang, X. | | 登録日 | 2011-08-24 | | 公開日 | 2011-11-30 | | 最終更新日 | 2011-12-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Discovery of potent and highly selective thienopyridine janus kinase 2 inhibitors.
J.Med.Chem., 54, 2011
|
|
6GIZ
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - SUBSTRATE COMPLEX | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | 登録日 | 2018-05-15 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GJ2
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - COMPLEX WITH INOSITOL HEXASULPHATE | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | 登録日 | 2018-05-15 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GIT
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - PRODUCT COMPLEX | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | 登録日 | 2018-05-15 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.418 Å) | | 主引用文献 | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GJA
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - H229A MUTANT | | 分子名称: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | 登録日 | 2018-05-16 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
7DPF
 
 | |
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | 登録日 | 2020-12-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | 分子名称: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
8HP5
 
 | | Crystal structure of (S)-2-haloacid dehalogenase | | 分子名称: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL | | 著者 | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | 登録日 | 2022-12-12 | | 公開日 | 2023-06-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
8HP7
 
 | | Crystal structure of (S)-2-haloacid dehalogenase K152A mutant trapped with (2R)-4-amino-2-hydroxybutanoic acid | | 分子名称: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, GAMMA-AMINO-BUTANOIC ACID | | 著者 | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | 登録日 | 2022-12-12 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
8HP6
 
 | | Crystal structure of (S)-2-haloacid dehalogenase D12A mutant | | 分子名称: | (S)-2-haloacid dehalogenase, SODIUM ION | | 著者 | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | 登録日 | 2022-12-12 | | 公開日 | 2023-06-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
6I9B
 
 | | NMR structure of the La module from human LARP4A | | 分子名称: | La-related protein 4 | | 著者 | Conte, M.R, Martino, L, Atkinson, R.A, Kelly, G, Cruz-Gallardo, I, De Tito, S, Trotta, R. | | 登録日 | 2018-11-22 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | LARP4A recognizes polyA RNA via a novel binding mechanism mediated by disordered regions and involving the PAM2w motif, revealing interplay between PABP, LARP4A and mRNA.
Nucleic Acids Res., 47, 2019
|
|
6JN2
 
 | | Crystal structure of the coiled-coil domains of human DOT1L in complex with AF10 | | 分子名称: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, Protein AF-10 | | 著者 | Song, X, Wang, M, Yang, N, Xu, R.M. | | 登録日 | 2019-03-13 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | A higher-order configuration of the heterodimeric DOT1L-AF10 coiled-coil domains potentiates their leukemogenenic activity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6A0Z
 
 | | Crystal structure of broadly neutralizing antibody 13D4 bound to H5N1 influenza hemagglutinin, HA head region | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 13D4, Fab Heavy Chain, ... | | 著者 | Li, S, Li, T. | | 登録日 | 2018-06-06 | | 公開日 | 2018-06-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.329 Å) | | 主引用文献 | Structural Basis for the Broad, Antibody-Mediated Neutralization of H5N1 Influenza Virus.
J. Virol., 92, 2018
|
|
5ZVV
 
 | | Structure of SeMet-phAimR | | 分子名称: | AimR transcriptional regulator, GLYCEROL | | 著者 | Cheng, W, Dou, C. | | 登録日 | 2018-05-13 | | 公開日 | 2018-09-05 | | 最終更新日 | 2019-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZW6
 
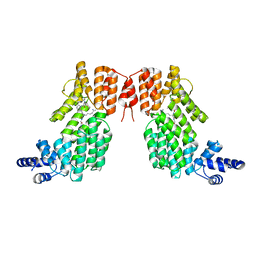 | | Structure of spAimR | | 分子名称: | AimR transcriptional regulator, GLY-MET-PRO-ARG-GLY-ALA | | 著者 | Cheng, W, Dou, C. | | 登録日 | 2018-05-14 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
6A0X
 
 | |
5ZW5
 
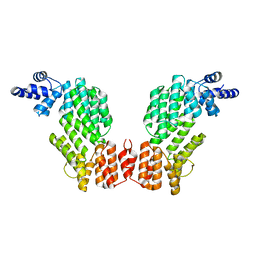 | | Structure of SeMet-spAimR | | 分子名称: | AimR transcriptional regulator | | 著者 | Cheng, W, Dou, C. | | 登録日 | 2018-05-14 | | 公開日 | 2018-08-29 | | 最終更新日 | 2019-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZVW
 
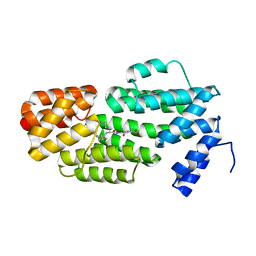 | | Structure of phAimR-Ligand | | 分子名称: | AimR transcriptional regulator, SER-ALA-ILE-ARG-GLY-ALA | | 著者 | Cheng, W, Dou, C. | | 登録日 | 2018-05-13 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.292 Å) | | 主引用文献 | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
6AJ2
 
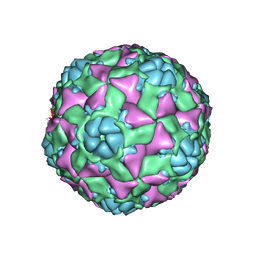 | | The structure of ICAM-5 triggered Enterovirus D68 virus A-particle | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | 著者 | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | 登録日 | 2018-08-26 | | 公開日 | 2018-11-07 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ0
 
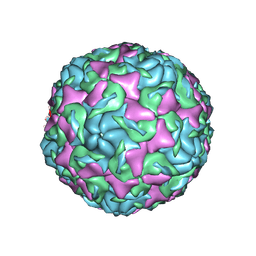 | | The structure of Enterovirus D68 mature virion | | 分子名称: | Capsid protein VP3, Capsid protein VP4, Viral protein 1, ... | | 著者 | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | 登録日 | 2018-08-25 | | 公開日 | 2018-11-07 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
7FJ1
 
 | | Cryo-EM structure of pseudorabies virus C-capsid | | 分子名称: | Capsid vertex component 1, DNA packaging tegument protein UL25, Major capsid protein, ... | | 著者 | Zheng, Q, Li, S, Zha, Z, Sun, H. | | 登録日 | 2021-08-02 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (4.43 Å) | | 主引用文献 | Structures of pseudorabies virus capsids.
Nat Commun, 13, 2022
|
|
