4JWO
 
 | | The crystal structure of a possible phosphate binding protein from Planctomyces limnophilus DSM 3776 | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | 著者 | Tan, K, Gu, M, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-03-27 | | 公開日 | 2013-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.601 Å) | | 主引用文献 | The crystal structure of a possible phosphate binding protein from Planctomyces limnophilus DSM 3776
To be Published
|
|
4MJM
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Short Internal Deletion of CBS Domain from Bacillus anthracis str. Ames | | 分子名称: | 1,2-ETHANEDIOL, Inosine-5'-monophosphate dehydrogenase | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-03 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2544 Å) | | 主引用文献 | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Short Internal Deletion of CBS Domain from Bacillus anthracis str. Ames
To be Published, 2013
|
|
4MYX
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ame complexed with P32 | | 分子名称: | 1,2-ETHANEDIOL, 2-chloro-5-{[(2-{3-[(1E)-N-hydroxyethanimidoyl]phenyl}propan-2-yl)carbamoyl]amino}benzamide, FORMIC ACID, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-28 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ame complexed with P32
To be Published
|
|
4MYA
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor A110 | | 分子名称: | 4-{(1R)-1-[1-(4-chlorophenyl)-1H-1,2,3-triazol-4-yl]ethoxy}quinolin-2(1H)-one, GLYCEROL, INOSINIC ACID, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-27 | | 公開日 | 2014-01-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8997 Å) | | 主引用文献 | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor A110
To be Published
|
|
4MY9
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor C91 | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, MALONATE ION, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-27 | | 公開日 | 2014-06-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5893 Å) | | 主引用文献 | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor C91
To be Published
|
|
4MY1
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ames complexed with P68 | | 分子名称: | 1-(4-bromophenyl)-3-(2-{3-[(1E)-N-hydroxyethanimidoyl]phenyl}propan-2-yl)urea, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-26 | | 公開日 | 2014-01-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5997 Å) | | 主引用文献 | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ames complexed with P68
To be Published
|
|
4MZ1
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12 | | 分子名称: | 1-(4-bromophenyl)-3-{2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}urea, ACETIC ACID, INOSINIC ACID, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-28 | | 公開日 | 2014-01-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3991 Å) | | 主引用文献 | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12
To be Published, 2013
|
|
4MZ8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with an Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91 | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-29 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5004 Å) | | 主引用文献 | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91
To be Published
|
|
3B85
 
 | |
5JMB
 
 | |
5JMU
 
 | | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 | | 分子名称: | ACETATE ION, MAGNESIUM ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | 著者 | Tan, K, Gu, M, Clancy, S, Joachimiak, A. | | 登録日 | 2016-04-29 | | 公開日 | 2016-06-29 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 (CASP target)
To Be Published
|
|
3C2Q
 
 | | Crystal structure of conserved putative LOR/SDH protein from Methanococcus maripaludis S2 | | 分子名称: | IMIDAZOLE, NICKEL (II) ION, Uncharacterized conserved protein | | 著者 | Duke, N, Gu, M, Mulligan, R, Conrad, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-01-25 | | 公開日 | 2008-02-05 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of conserved putative LOR/SDH protein from Methanococcus maripaludis S2
To be Published
|
|
3BY6
 
 | |
3C7J
 
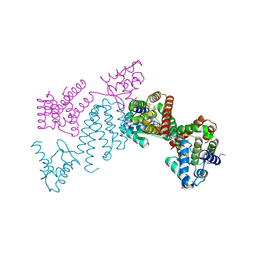 | | Crystal structure of transcriptional regulator (GntR family member) from Pseudomonas syringae pv. tomato str. DC3000 | | 分子名称: | NICKEL (II) ION, Transcriptional regulator, GntR family | | 著者 | Nocek, B, Sather, A, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-02-07 | | 公開日 | 2008-02-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of transcriptional regulator (GntR family member) from Pseudomonas syringae pv. tomato str. DC3000.
To be Published
|
|
3BRO
 
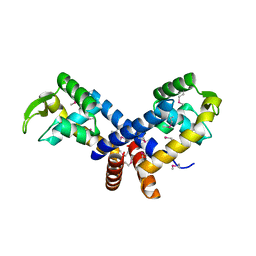 | | Crystal structure of the transcription regulator MarR from Oenococcus oeni PSU-1 | | 分子名称: | CHLORIDE ION, GLYCEROL, Transcriptional regulator | | 著者 | Kim, Y, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-12-21 | | 公開日 | 2008-01-15 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Crystal Structure of the Transcription Regulator MarR from Oenococcus oeni PSU-1.
To be Published
|
|
3CDL
 
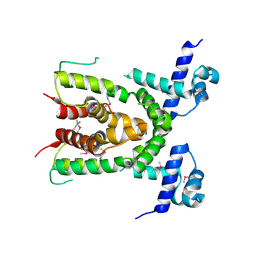 | |
3D7L
 
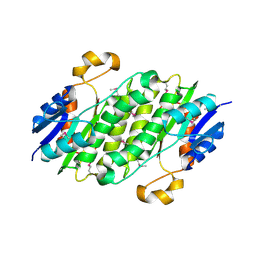 | |
3DOB
 
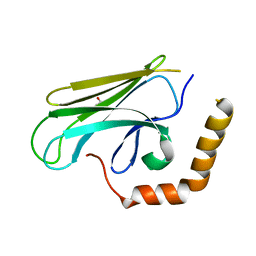 | | Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans. | | 分子名称: | BETA-MERCAPTOETHANOL, Heat shock 70 kDa protein F44E5.5 | | 著者 | Osipiuk, J, Hatzos, C, Gu, M, Zhang, R, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-07-03 | | 公開日 | 2008-07-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | X-ray crystal structure of Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans.
To be Published
|
|
3DQG
 
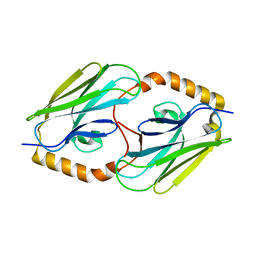 | | Peptide-binding domain of heat shock 70 kDa protein F, mitochondrial precursor, from Caenorhabditis elegans. | | 分子名称: | Heat shock 70 kDa protein F | | 著者 | Osipiuk, J, Mulligan, R, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-07-09 | | 公開日 | 2008-07-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | X-ray crystal structure of peptide-binding domain of heat shock 70 kDa protein F, mitochondrial precursor, from Caenorhabditis elegans.
To be Published
|
|
7VFJ
 
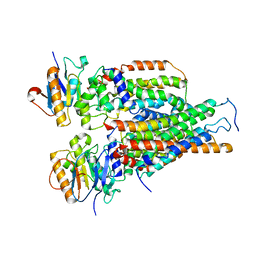 | | Cytochrome c-type biogenesis protein CcmABCD | | 分子名称: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | 著者 | Zhu, J.P, Zhang, K, Li, J, Zheng, W, Gu, M. | | 登録日 | 2021-09-13 | | 公開日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (3.98 Å) | | 主引用文献 | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7VFP
 
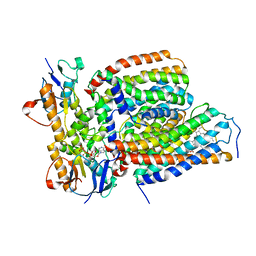 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with heme and single ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | 著者 | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | 登録日 | 2021-09-13 | | 公開日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (4.03 Å) | | 主引用文献 | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
4PZ0
 
 | | The crystal structure of a solute binding protein from Bacillus anthracis str. Ames in complex with quorum-sensing signal autoinducer-2 (AI-2) | | 分子名称: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-03-28 | | 公開日 | 2014-04-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | The crystal structure of a solute binding protein from Bacillus anthracis str. Ames in complex with quorum-sensing signal autoinducer-2 (AI-2).
To be Published
|
|
4RD7
 
 | | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205 | | 分子名称: | Cupin 2 conserved barrel domain protein, GLYCEROL, SULFATE ION | | 著者 | Tan, K, Gu, M, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-09-18 | | 公開日 | 2014-10-01 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.571 Å) | | 主引用文献 | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205
To be Published
|
|
6NRU
 
 | | Crystal Structure of the Alpha-ribazole Phosphatase from Shigella flexneri | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, CobC, ... | | 著者 | Kim, Y, Gu, M, Shatsman, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-01-24 | | 公開日 | 2019-03-06 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (2.505 Å) | | 主引用文献 | Crystal Structure of the Alpha-ribazole Phosphatase from Shigella flexneri
To Be Published
|
|
2NR7
 
 | | Structural Genomics, the crystal structure of putative secretion activator protein from Porphyromonas gingivalis W83 | | 分子名称: | Secretion activator protein, putative | | 著者 | Tan, K, Bigelow, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-11-01 | | 公開日 | 2006-12-05 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | The crystal structure of putative secretion activator protein from Porphyromonas gingivalis W83
To be Published
|
|
