3T6B
 
 | |
5M58
 
 | |
4CE5
 
 | | First crystal structure of an (R)-selective omega-transaminase from Aspergillus terreus | | 分子名称: | AT-OMEGATA, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Lyskowski, A, Gruber, C, Steinkellner, G, Schurmann, M, Schwab, H, Gruber, K, Steiner, K. | | 登録日 | 2013-11-08 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Crystal Structure of an (R)-Selective Omega-Transaminase from Aspergillus Terreus
Plos One, 9, 2014
|
|
3ZOF
 
 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound benzene-1,4-diol | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, benzene-1,4-diol | | 著者 | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | 登録日 | 2013-02-21 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOG
 
 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound 1-Cyclohex-2-enone | | 分子名称: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, cyclohex-2-en-1-one | | 著者 | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | 登録日 | 2013-02-21 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOE
 
 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound p-hydroxybenzaldehyde | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, P-HYDROXYBENZALDEHYDE | | 著者 | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | 登録日 | 2013-02-21 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOD
 
 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound benzene-1,4-diol | | 分子名称: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, benzene-1,4-diol | | 著者 | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | 登録日 | 2013-02-21 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOH
 
 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound 1-Cyclohex-2-enone | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, cyclohex-2-en-1-one | | 著者 | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | 登録日 | 2013-02-21 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOC
 
 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound p-hydroxybenzaldehyde | | 分子名称: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, P-HYDROXYBENZALDEHYDE | | 著者 | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | 登録日 | 2013-02-21 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
8B30
 
 | |
8C66
 
 | |
2AN7
 
 | | Solution structure of the bacterial antidote ParD | | 分子名称: | Protein parD | | 著者 | Oberer, M, Zangger, K, Gruber, K, Keller, W. | | 登録日 | 2005-08-11 | | 公開日 | 2006-09-05 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of ParD, the antidote of the ParDE toxin antitoxin module, provides the structural basis for DNA and toxin binding.
Protein Sci., 16, 2007
|
|
8PUN
 
 | |
6EZD
 
 | |
8OIM
 
 | |
5E4D
 
 | |
5E4B
 
 | |
5E4M
 
 | |
4BIF
 
 | | Biochemical and structural characterisation of a novel manganese- dependent hydroxynitrile lyase from bacteria | | 分子名称: | CUPIN 2 CONSERVED BARREL DOMAIN PROTEIN, MANGANESE (II) ION | | 著者 | Hajnal, I, Lyskowski, A, Hanefeld, U, Gruber, K, Schwab, H, Steiner, K. | | 登録日 | 2013-04-10 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Biochemical and Structural Characterisation of a Novel Bacterial Manganese-Dependent Hydroxynitrile Lyase.
FEBS J., 280, 2013
|
|
5E46
 
 | |
7A0Q
 
 | |
7A0T
 
 | |
7LQU
 
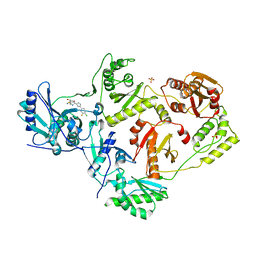 | | Crystal Structure of HIV-1 RT in Complex with NBD-14075 | | 分子名称: | Reverse transcriptase p51, Reverse transcriptase p66, SULFATE ION, ... | | 著者 | Losada, N, Ruiz, F.X, Gruber, K, Das, K, Arnold, E. | | 登録日 | 2021-02-15 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | HIV-1 gp120 Antagonists Also Inhibit HIV-1 Reverse Transcriptase by Bridging the NNRTI and NRTI Sites.
J.Med.Chem., 64, 2021
|
|
8AWN
 
 | | Crystal structure of a manganese-containing cupin (tm1459) from Thermotoga maritima, variant C106Q | | 分子名称: | CHLORIDE ION, Cupin_2 domain-containing protein | | 著者 | Grininger, C, Steiner, K, Gruber, K, Pavkov-Keller, T. | | 登録日 | 2022-08-30 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Engineering TM1459 for Stabilisation against Inactivation by Amino Acid Oxidation
Chem Ing Tech, 2023
|
|
8AWP
 
 | |
