7LQU
 
 | | Crystal Structure of HIV-1 RT in Complex with NBD-14075 | | Descriptor: | Reverse transcriptase p51, Reverse transcriptase p66, SULFATE ION, ... | | Authors: | Losada, N, Ruiz, F.X, Gruber, K, Das, K, Arnold, E. | | Deposit date: | 2021-02-15 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | HIV-1 gp120 Antagonists Also Inhibit HIV-1 Reverse Transcriptase by Bridging the NNRTI and NRTI Sites.
J.Med.Chem., 64, 2021
|
|
7A0Q
 
 | |
7A0T
 
 | |
4ZXF
 
 | | Crystal Structure of a Soluble Variant of Monoglyceride Lipase from Saccharomyces Cerevisiae in Complex with a Substrate Analog | | Descriptor: | 1-{3-[(R)-hydroxy(octadecyloxy)phosphoryl]propyl}triaza-1,2-dien-2-ium, Monoglyceride lipase, NITRATE ION, ... | | Authors: | Aschauer, P, Lichtenegger, J, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-20 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
4ZWN
 
 | | Crystal Structure of a Soluble Variant of the Monoglyceride Lipase from Saccharomyces Cerevisiae | | Descriptor: | Monoglyceride lipase, NITRATE ION, SODIUM ION, ... | | Authors: | Aschauer, P, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-19 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
8PUN
 
 | |
4BIF
 
 | | Biochemical and structural characterisation of a novel manganese- dependent hydroxynitrile lyase from bacteria | | Descriptor: | CUPIN 2 CONSERVED BARREL DOMAIN PROTEIN, MANGANESE (II) ION | | Authors: | Hajnal, I, Lyskowski, A, Hanefeld, U, Gruber, K, Schwab, H, Steiner, K. | | Deposit date: | 2013-04-10 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel Bacterial Manganese-Dependent Hydroxynitrile Lyase.
FEBS J., 280, 2013
|
|
3FW7
 
 | |
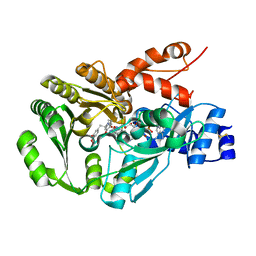
3FWA
 
 | |
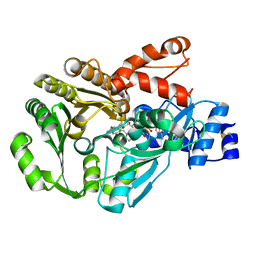
3FW8
 
 | |
2XAA
 
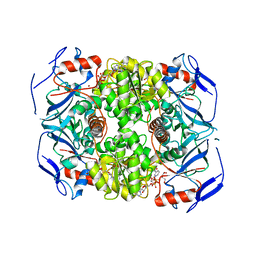 | | Alcohol dehydrogenase ADH-'A' from Rhodococcus ruber DSM 44541 at pH 8.5 in complex with NAD and butane-1,4-diol | | Descriptor: | 1,4-BUTANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SECONDARY ALCOHOL DEHYDROGENASE, ... | | Authors: | Kroutil, W, Gruber, K, Grogan, G. | | Deposit date: | 2010-03-30 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into Substrate Specificity and Solvent Tolerance in Alcohol Dehydrogenase Adh-'A' from Rhodococcus Ruber Dsm 44541.
Chem.Commun.(Camb.), 46, 2010
|
|
3FW9
 
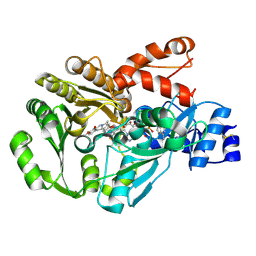 | | Structure of berberine bridge enzyme in complex with (S)-scoulerine | | Descriptor: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winkler, A, Macheroux, P, Gruber, K. | | Deposit date: | 2009-01-17 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structural roles of biocovalent flaninylation in berberine bridge enzyme
to be published
|
|
4V15
 
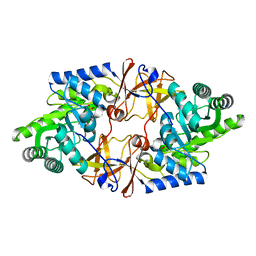 | | Crystal structure of D-threonine aldolase from Alcaligenes xylosoxidans | | Descriptor: | D-THREONINE ALDOLASE, MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Uhl, M.K, Oberdorfer, G, Steinkellner, G, Riegler, L, Schuermann, M, Gruber, K. | | Deposit date: | 2014-09-24 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of D-Threonine Aldolase from Alcaligenes Xylosoxidans Provides Insight Into a Metal Ion Assisted Plp-Dependent Mechanism.
Plos One, 10, 2015
|
|
4WJL
 
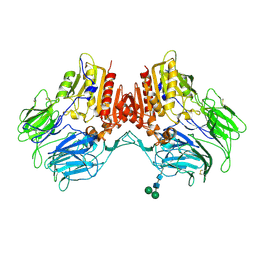 | | Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Inactive dipeptidyl peptidase 10, ... | | Authors: | Bezerra, G.A, Dobrovetsky, E, Seitova, A, Fedosyuk, S, Dhe-Paganon, S, Gruber, K. | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels.
Sci Rep, 5, 2015
|
|
8OIM
 
 | |
8P7E
 
 | |
5NY5
 
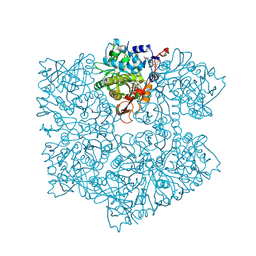 | | The apo structure of 3,4-dihydroxybenzoic acid decarboxylases from Enterobacter cloacae | | Descriptor: | 3,4-dihydroxybenzoate decarboxylase, GLYCEROL | | Authors: | Dordic, A, Gruber, K, Payer, S, Glueck, S, Pavkov-Keller, T, Marshall, S, Leys, D. | | Deposit date: | 2017-05-11 | | Release date: | 2017-09-13 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8P8F
 
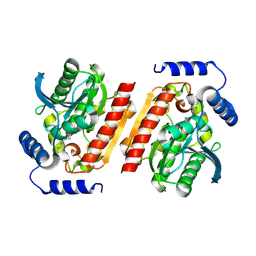 | |
6GHW
 
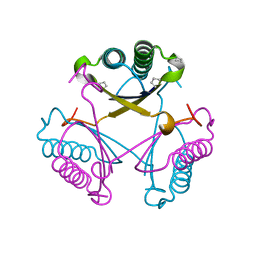 | | Substituting the prolines of 4-oxalocrotonate tautomerase with non-canonical analogue (2S)-3,4-dehydroproline | | Descriptor: | 2-hydroxymuconate tautomerase, CALCIUM ION | | Authors: | Pavkov-Keller, T, Lukesch, M.S, Wiltschi, B, Gruber, K. | | Deposit date: | 2018-05-09 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substituting the catalytic proline of 4-oxalocrotonate tautomerase with non-canonical analogues reveals a finely tuned catalytic system.
Sci Rep, 9, 2019
|
|
1JU2
 
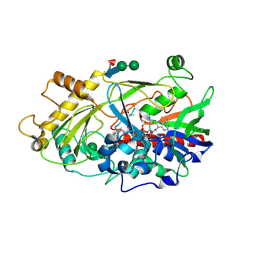 | | Crystal structure of the hydroxynitrile lyase from almond | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dreveny, I, Gruber, K, Glieder, A, Thompson, A, Kratky, C. | | Deposit date: | 2001-08-23 | | Release date: | 2002-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The hydroxynitrile lyase from almond: a lyase that looks like an oxidoreductase.
Structure, 9, 2001
|
|
4U36
 
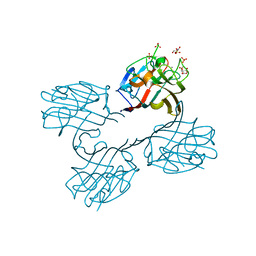 | | Crystal structure of a seed lectin from Vatairea macrocarpa complexed with Tn-antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, CITRIC ACID, ... | | Authors: | Sousa, B.L, Silva-Filho, J.C, Kumar, P, Lyskowski, A, Bezerra, G.A, Delatorre, P, Rocha, B.A.M, Nagano, C.S, Gruber, K, Cavada, B.S. | | Deposit date: | 2014-07-18 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structure of a new Tn antigen-binding lectin from Vatairea macrocarpa and a comparative analysis of Tn-binding legume lectins.
Int.J.Biochem.Cell Biol., 59C, 2014
|
|
4U2A
 
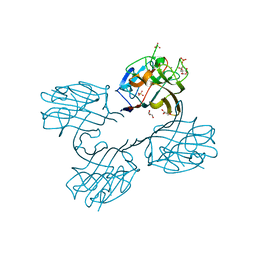 | | Structure of a lectin from the seeds of Vatairea macrocarpa complexed with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, CITRIC ACID, ... | | Authors: | Sousa, B.L, Silva-Filho, J.C, Kumar, P, Lyskowski, A, Bezerra, G.A, Delatorre, P, Rocha, B.A.M, Nagano, C.S, Gruber, K, Cavada, B.S. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | High-resolution structure of a new Tn antigen-binding lectin from Vatairea macrocarpa and a comparative analysis of Tn-binding legume lectins.
Int.J.Biochem.Cell Biol., 59C, 2014
|
|
4UD8
 
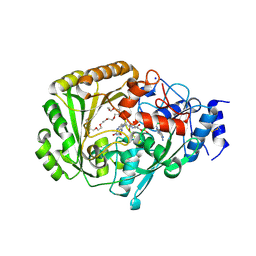 | | AtBBE15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Daniel, B, Steiner, B, Pavkov-Keller, T, Dordic, A, Gutmann, A, Sensen, C.W, Nidetzky, B, van der Graaff, E, Wallner, S, Gruber, K, Macheroux, P. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Oxidation of Monolignols by Members of the Berberine Bridge Enzyme Family Suggests a Role in Cell Wall Metabolism.
J.Biol.Chem., 290, 2015
|
|
3GDN
 
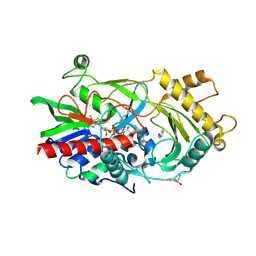 | | Almond hydroxynitrile lyase in complex with benzaldehyde | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dreveny, I, Gruber, K, Kratky, C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Substrate binding in the FAD-dependent hydroxynitrile lyase from almond provides insight into the mechanism of cyanohydrin formation and explains the absence of dehydrogenation activity.
Biochemistry, 48, 2009
|
|
5A2G
 
 | | An esterase from anaerobic Clostridium hathewayi can hydrolyze aliphatic aromatic polyesters | | Descriptor: | CARBOXYLIC ESTER HYDROLASE, PHOSPHATE ION | | Authors: | Hromic, A, Pavkov Keller, T, Steinkellner, G, Gruber, K, Perz, V, Baumschlager, A, Bleymaier, K, Zitzenbacher, S, Zankel, A, Mayrhofer, C, Sinkel, C, Kueper, U, Schlegel, K.A, Ribitsch, D, Guebitz, G.M. | | Deposit date: | 2015-05-19 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | An Esterase from Anaerobic Clostridium Hathewayi Can Hydrolyze Aliphatic-Aromatic Polyesters.
Environ.Sci.Tech., 50, 2016
|
|
