6WLE
 
 | | Crystal structure of the Zeitlupe light-state mimic G46A | | Descriptor: | 1,2-ETHANEDIOL, Adagio protein 1, FLAVIN MONONUCLEOTIDE | | Authors: | Zoltowski, B, Green, R. | | Deposit date: | 2020-04-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Steric and Electronic Interactions at Gln154 in ZEITLUPE Induce Reorganization of the LOV Domain Dimer Interface.
Biochemistry, 60, 2021
|
|
6WLP
 
 | | Crystal Structure of the ZTL light-state mimic G46S | | Descriptor: | 1,2-ETHANEDIOL, Adagio protein 1, FLAVIN MONONUCLEOTIDE | | Authors: | Zoltowski, B, Green, R. | | Deposit date: | 2020-04-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Steric and Electronic Interactions at Gln154 in ZEITLUPE Induce Reorganization of the LOV Domain Dimer Interface.
Biochemistry, 60, 2021
|
|
3U7G
 
 | | Crystal structure of mPNKP catalytic fragment (D170A) bound to single-stranded DNA (TCCTAp) | | Descriptor: | Bifunctional polynucleotide phosphatase/kinase, DNA, GLYCEROL, ... | | Authors: | Coquelle, N, Havali, Z, Bernstein, N, Green, R, Glover, J.N.M. | | Deposit date: | 2011-10-13 | | Release date: | 2011-12-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the phosphatase activity of polynucleotide kinase/phosphatase on single- and double-stranded DNA substrates.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U7E
 
 | | Crystal structure of mPNKP catalytic fragment (D170A) | | Descriptor: | Bifunctional polynucleotide phosphatase/kinase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Coquelle, N, Havali, Z, Bernstein, N, Green, R, Glover, J.N.M. | | Deposit date: | 2011-10-13 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the phosphatase activity of polynucleotide kinase/phosphatase on single- and double-stranded DNA substrates.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U7F
 
 | | Crystal structure of mPNKP catalytic fragment (D170A) bound to single-stranded DNA (TCCTCp) | | Descriptor: | Bifunctional polynucleotide phosphatase/kinase, DNA, GLYCEROL, ... | | Authors: | Coquelle, N, Havali, Z, Bernstein, N, Green, R, Glover, J.N.M. | | Deposit date: | 2011-10-13 | | Release date: | 2011-12-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the phosphatase activity of polynucleotide kinase/phosphatase on single- and double-stranded DNA substrates.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U7H
 
 | | Crystal structure of mPNKP catalytic fragment (D170A) bound to single-stranded DNA (TCCTTp) | | Descriptor: | Bifunctional polynucleotide phosphatase/kinase, DNA, GLYCEROL, ... | | Authors: | Coquelle, N, Havali, Z, Bernstein, N, Green, R, Glover, J.N.M. | | Deposit date: | 2011-10-13 | | Release date: | 2011-12-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the phosphatase activity of polynucleotide kinase/phosphatase on single- and double-stranded DNA substrates.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1JNX
 
 | |
1YJ5
 
 | | Molecular architecture of mammalian polynucleotide kinase, a DNA repair enzyme | | Descriptor: | 5' polynucleotide kinase-3' phosphatase FHA domain, 5' polynucleotide kinase-3' phosphatase catalytic domain, SULFATE ION | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-13 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
1YJM
 
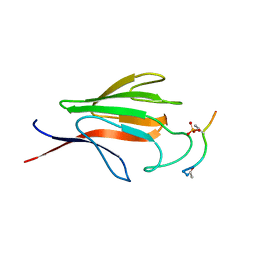 | | Crystal structure of the FHA domain of mouse polynucleotide kinase in complex with an XRCC4-derived phosphopeptide. | | Descriptor: | 12-mer peptide from DNA-repair protein XRCC4, Polynucleotide 5'-hydroxyl-kinase | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
6T7I
 
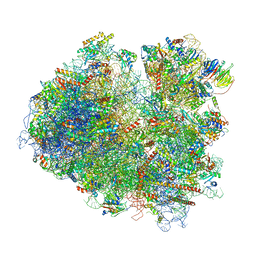 | | Structure of yeast 80S ribosome stalled on the CGA-CGA inhibitory codon combination. | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-22 | | Release date: | 2019-12-25 | | Last modified: | 2020-02-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6T4Q
 
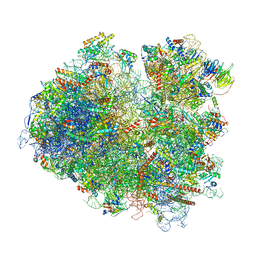 | | Structure of yeast 80S ribosome stalled on the CGA-CCG inhibitory codon combination. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-14 | | Release date: | 2019-12-25 | | Last modified: | 2020-02-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6T83
 
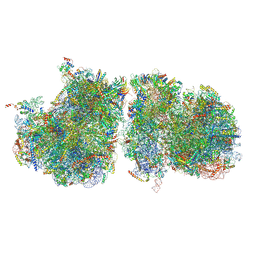 | | Structure of yeast disome (di-ribosome) stalled on poly(A) tract. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-24 | | Release date: | 2019-12-25 | | Last modified: | 2020-02-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6T7T
 
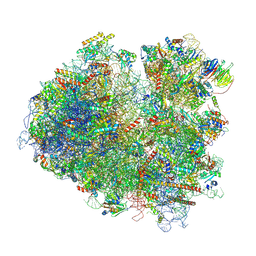 | | Structure of yeast 80S ribosome stalled on poly(A) tract. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-23 | | Release date: | 2019-12-25 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
1TIB
 
 | |
7QGH
 
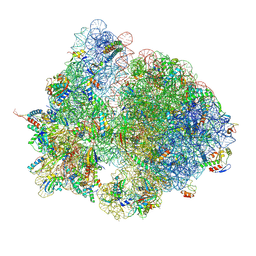 | | Structure of the E. coli disome - collided 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.48 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QG8
 
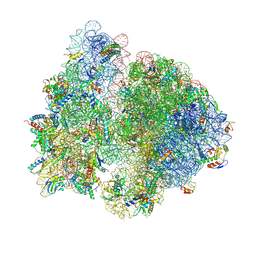 | | Structure of the collided E. coli disome - VemP-stalled 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-07 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QGU
 
 | | Structure of the B. subtilis disome - stalled 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QH4
 
 | | Structure of the B. subtilis disome - collided 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (5.45 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QGN
 
 | | Structure of the SmrB-bound E. coli disome - stalled 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-09 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QGR
 
 | | Structure of the SmrB-bound E. coli disome - collided 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-09 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QQ3
 
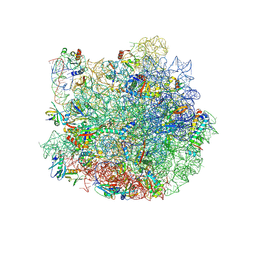 | | Cryo-EM structure of the E.coli 50S ribosomal subunit in complex with the antibiotic Myxovalargin A. | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Koller, T.O, Beckert, B, Wilson, D.N. | | Deposit date: | 2022-01-06 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | The Myxobacterial Antibiotic Myxovalargin: Biosynthesis, Structural Revision, Total Synthesis, and Molecular Characterization of Ribosomal Inhibition.
J.Am.Chem.Soc., 145, 2023
|
|
1E0W
 
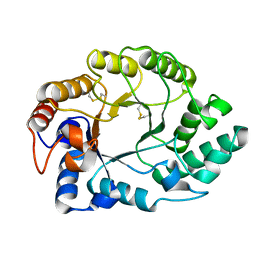 | | Xylanase 10A from Sreptomyces lividans. native structure at 1.2 angstrom resolution | | Descriptor: | ENDO-1,4-BETA-XYLANASE A | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1E0X
 
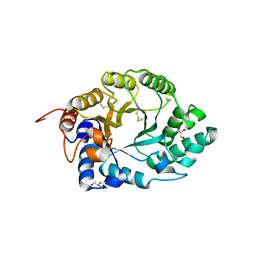 | | XYLANASE 10A FROM SREPTOMYCES LIVIDANS. XYLOBIOSYL-ENZYME INTERMEDIATE AT 1.65 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1E0V
 
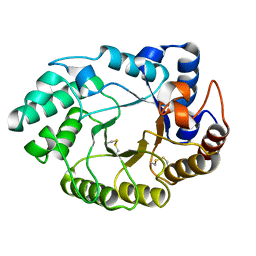 | | Xylanase 10A from Sreptomyces lividans. cellobiosyl-enzyme intermediate at 1.7 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
6ZVI
 
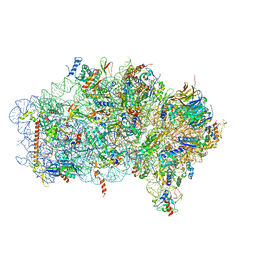 | | Mbf1-ribosome complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S10-A, ... | | Authors: | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
