3J9S
 
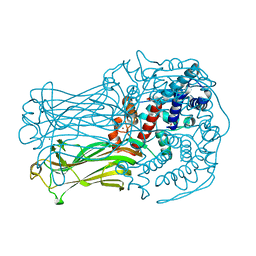 | |
5FXH
 
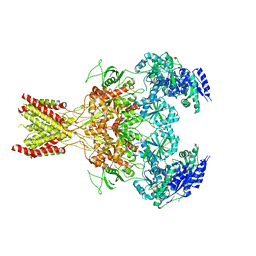 | | GluN1b-GluN2B NMDA receptor in non-active-1 conformation | | 分子名称: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | 著者 | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | 登録日 | 2016-03-02 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
9C8V
 
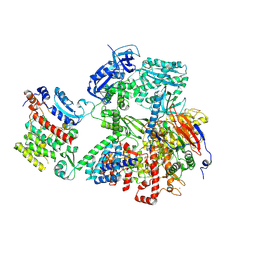 | | Human DNA polymerase alpha/primase - CHAPSO (4 mM) | | 分子名称: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase large subunit, ... | | 著者 | Abe, K.M, Li, G, He, Q, Grant, T, Lim, C. | | 登録日 | 2024-06-13 | | 公開日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Human DNA polymerase alpha/primase - CHAPSO (4 mM)
To Be Published
|
|
9C8U
 
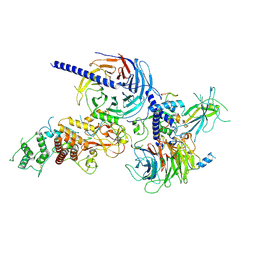 | | Human PRC2 - RvLEAM (short) (1:6 molar ratio), cross-linked 10 min | | 分子名称: | Isoform 2 of Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, Polycomb protein SUZ12, ... | | 著者 | Abe, K.M, Li, G, He, Q, Grant, T, Lim, C. | | 登録日 | 2024-06-13 | | 公開日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Human PRC2 - RvLEAM (short) (1:6 molar ratio), cross-linked 10 min
To Be Published
|
|
8DB3
 
 | | Crystal structure of KaiC with truncated C-terminal coiled-coil domain | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiC | | 著者 | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | 登録日 | 2022-06-14 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
8DBA
 
 | | Crystal structure of dodecameric KaiC | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiC, MAGNESIUM ION | | 著者 | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | 登録日 | 2022-06-14 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
6OJ3
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ4
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ5
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP_RNA) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ6
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP_RNA) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase, Template, ... | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
8FMA
 
 | | Nodavirus RNA replication proto-crown, detergent-solubliized C11 multimer | | 分子名称: | RNA-directed RNA polymerase | | 著者 | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | 登録日 | 2022-12-22 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM9
 
 | | Nodavirus RNA replication proto-crown, detergent-solubliized C12 multimer | | 分子名称: | RNA-directed RNA polymerase | | 著者 | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | 登録日 | 2022-12-22 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FMB
 
 | | Nodavirus RNA replication protein A polymerase domain, local refinement | | 分子名称: | RNA-directed RNA polymerase | | 著者 | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | 登録日 | 2022-12-22 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8VY3
 
 | | Human DNA polymerase alpha/primase - AavLEA1 (1:40 molar ratio) | | 分子名称: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase large subunit, ... | | 著者 | Abe, K.M, Li, G, Grant, T, Lim, C.J. | | 登録日 | 2024-02-06 | | 公開日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Human PRC2 - AavLEA1 (1:40 molar ratio)
To Be Published
|
|
8VT0
 
 | | SPOT-RASTR - a cryo-EM specimen preparation technique that overcomes problems with preferred orientation and the air/water interface | | 分子名称: | Beta-galactosidase, MAGNESIUM ION | | 著者 | Esfahani, B.G, Randolph, P, Peng, R, Grant, T, Stroupe, M.E, Stagg, S.M. | | 登録日 | 2024-01-25 | | 公開日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | SPOT-RASTR-A cryo-EM specimen preparation technique that overcomes problems with preferred orientation and the air/water interface.
Pnas Nexus, 3, 2024
|
|
2VY9
 
 | | Molecular architecture of the stressosome, a signal integration and transduction hub | | 分子名称: | ANTI-SIGMA-FACTOR ANTAGONIST | | 著者 | Marles-Wright, J, Grant, T, Delumeau, O, van Duinen, G, Firbank, S.J, Lewis, P.J, Murray, J.W, Newman, J.A, Quin, M.B, Race, P.R, Rohou, A, Tichelaar, W, van Heel, M, Lewis, R.J. | | 登録日 | 2008-07-21 | | 公開日 | 2008-10-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular Architecture of the "Stressosome," a Signal Integration and Transduction Hub
Science, 322, 2008
|
|
8FWI
 
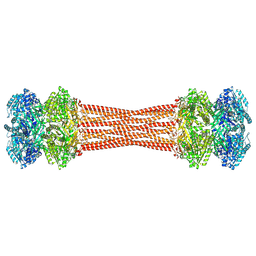 | | Structure of dodecameric KaiC-RS-S413E/S414E solved by cryo-EM | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiC, ... | | 著者 | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | 登録日 | 2023-01-22 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
8FWJ
 
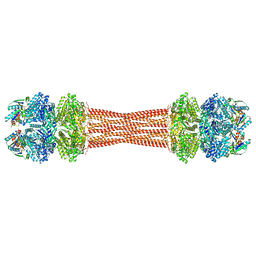 | | Structure of dodecameric KaiC-RS-S413E/S414E complexed with KaiB-RS solved by cryo-EM | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiB, ... | | 著者 | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | 登録日 | 2023-01-22 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
8FAK
 
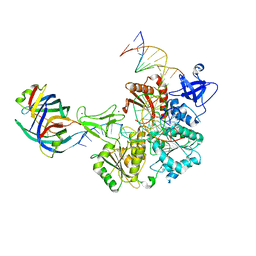 | | DNA replication fork binding triggers structural changes in the PriA DNA helicase that regulate the PriA-PriB replication restart pathway in E. coli | | 分子名称: | DNA (5'-D(P*CP*AP*GP*AP*CP*TP*CP*AP*TP*TP*TP*AP*GP*CP*CP*CP*TP*TP*AP*TP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*AP*TP*AP*AP*GP*GP*GP*CP*TP*GP*AP*GP*CP*AP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*GP*CP*GP*TP*GP*CP*TP*C)-3'), ... | | 著者 | Duckworth, A.T, Ducos, P.L, McMillan, S.D, Satyshur, K.A, Blumenthal, K.H, Deorio, H.R, Larson, J.A, Sandler, S.J, Grant, T, Keck, J.L. | | 登録日 | 2022-11-28 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Replication fork binding triggers structural changes in the PriA helicase that govern DNA replication restart in E. coli.
Nat Commun, 14, 2023
|
|
6N1R
 
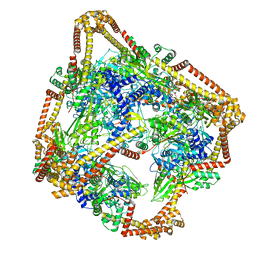 | | Tetrahedral oligomeric complex of GyrA N-terminal fragment, solved by cryoEM in tetrahedral symmetry | | 分子名称: | DNA gyrase subunit A | | 著者 | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | 登録日 | 2018-11-10 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
6CNA
 
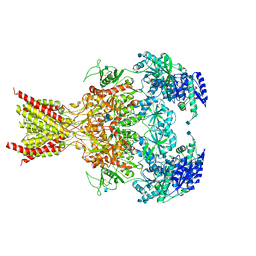 | | GluN1-GluN2B NMDA receptors with exon 5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | 著者 | Furukawa, H, Grant, T, Grigorieff, N. | | 登録日 | 2018-03-07 | | 公開日 | 2018-10-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural Mechanism of Functional Modulation by Gene Splicing in NMDA Receptors.
Neuron, 98, 2018
|
|
6N1P
 
 | | Dihedral oligomeric complex of GyrA N-terminal fragment with DNA, solved by cryoEM in C2 symmetry | | 分子名称: | DNA (44-MER), DNA gyrase subunit A | | 著者 | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | 登録日 | 2018-11-10 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (6.35 Å) | | 主引用文献 | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
6N1Q
 
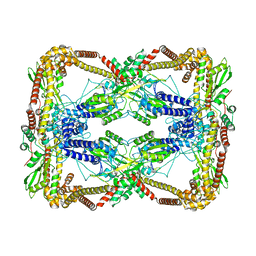 | | Dihedral oligomeric complex of GyrA N-terminal fragment, solved by cryoEM in D2 symmetry | | 分子名称: | DNA gyrase subunit A | | 著者 | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | 登録日 | 2018-11-10 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (5.16 Å) | | 主引用文献 | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
6OUC
 
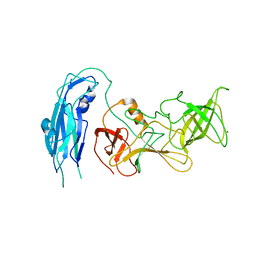 | | Asymmetric focsued reconstruction of human norovirus GII.2 Snow Mountain Virus strain VLP asymmetric unit in T=1 symmetry | | 分子名称: | Viral protein 1, ZINC ION | | 著者 | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | 登録日 | 2019-05-04 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OUU
 
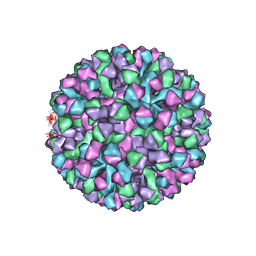 | | Symmetric reconstruction of human norovirus GII.4 Minerva strain VLP in T=4 symmetry | | 分子名称: | Major capsid protein | | 著者 | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | 登録日 | 2019-05-05 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
