6ZUS
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | DI(HYDROXYETHYL)ETHER, Extracellular protein 11-1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
6ZUQ
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | Extracellular protein 11-1, GLYCEROL, ZINC ION | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
7AD5
 
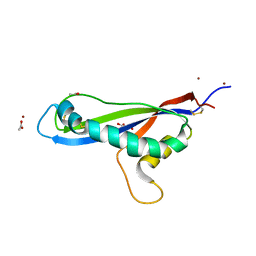 | | Crystal structure of the effector AvrLm5-9 from Leptosphaeria maculans | | Descriptor: | ACETATE ION, Avirulence protein LmJ1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-09-14 | | Release date: | 2021-10-06 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
7B76
 
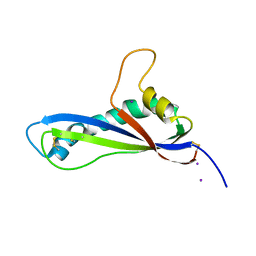 | | Crystal structure of the effector AvrLm5-9 from Leptosphaeria maculans | | Descriptor: | Avirulence protein LmJ1, IODIDE ION | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-03-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
6TWR
 
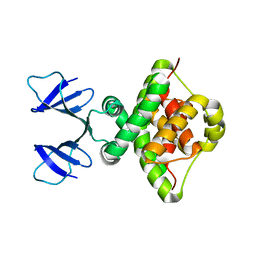 | |
5JHJ
 
 | | M. Oryzae effector AVR-Pia mutant H3 | | Descriptor: | Antivirulence protein AVR-Pia | | Authors: | Padilla, A, deGuillen, K. | | Deposit date: | 2016-04-21 | | Release date: | 2017-03-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Recognition of the Magnaporthe oryzae Effector AVR-Pia by the Decoy Domain of the Rice NLR Immune Receptor RGA5.
Plant Cell, 29, 2017
|
|
2IT7
 
 | |
2IT8
 
 | |
2MYW
 
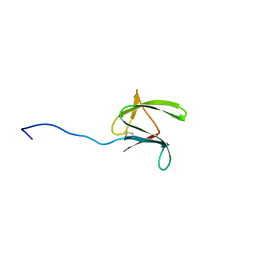 | |
2MYV
 
 | |
