6YEF
 
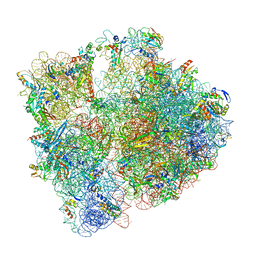 | | 70S initiation complex with assigned rRNA modifications from Staphylococcus aureus | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S11, ... | | Authors: | Fatkhullin, B, Golubev, A, Khusainov, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the ribosome functional complex of the human pathogen Staphylococcus aureus at 3.2 angstrom resolution.
Febs Lett., 594, 2020
|
|
6RK3
 
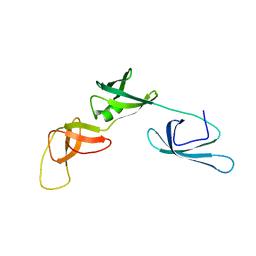 | | Solution structure of the ribosome Elongation Factor P (EF-P) from Staphylococcus aureus | | Descriptor: | Elongation factor P | | Authors: | Usachev, K, Fatkhullin, B, Gabdulkhakov, A, Khusainov, I, Golubev, A, Validov, S, Yusupova, G, Yusupov, M. | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR and crystallographic structural studies of the Elongation factor P from Staphylococcus aureus.
Eur.Biophys.J., 49, 2020
|
|
3ILN
 
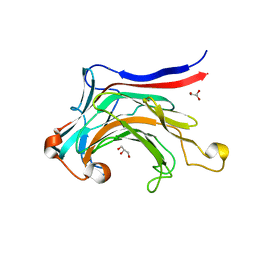 | | X-ray structure of the laminarinase from Rhodothermus marinus | | Descriptor: | CALCIUM ION, GLYCEROL, Laminarinase | | Authors: | Bleicher, L, Golubev, A, Rojas, A.L, Nascimento, A.S, Polikarpov, I. | | Deposit date: | 2009-08-07 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis of the thermostability and thermophilicity of laminarinases: X-ray structure of the hyperthermostable laminarinase from Rhodothermus marinus and molecular dynamics simulations.
J.Phys.Chem.B, 115, 2011
|
|
6RJI
 
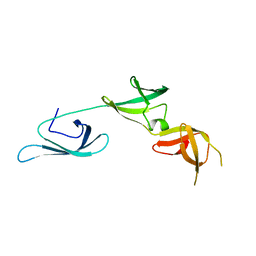 | | X-ray structure of the elongation factor P of S. aureus | | Descriptor: | Elongation factor P | | Authors: | Fatkhullin, B.F, Golubev, A.A, Gabdulkhakov, A.G, Khusainov, I.S, Validov, S.Z, Usachev, K.S, Yusupova, G, Yusupov, M.M. | | Deposit date: | 2019-04-27 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | NMR and crystallographic structural studies of the Elongation factor P from Staphylococcus aureus.
Eur.Biophys.J., 49, 2020
|
|
4UB7
 
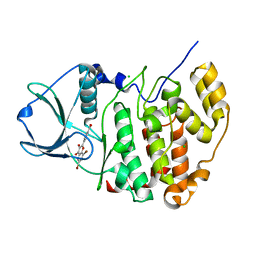 | | High-salt structure of protein kinase CK2 catalytic subunit with 4'-carboxy-6,8-bromo-flavonol (FLC26) showing an extreme distortion of the ATP-binding loop combined with a pi-halogen bond | | Descriptor: | 4-(6,8-dibromo-3-hydroxy-4-oxo-4H-chromen-2-yl)benzoic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Niefind, K, Bischoff, N, Guerra, B, Golub, A, Issinger, O.-G. | | Deposit date: | 2014-08-12 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Note of Caution on the Role of Halogen Bonds for Protein Kinase/Inhibitor Recognition Suggested by High- And Low-Salt CK2 alpha Complex Structures.
Acs Chem.Biol., 10, 2015
|
|
4UBA
 
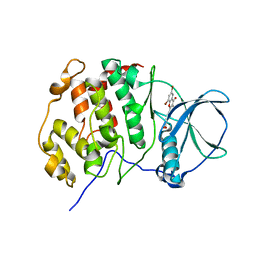 | | Low-salt structure of protein kinase CK2 catalytic subunit with 4'-carboxy-6,8-bromo-flavonol (FLC26) | | Descriptor: | 4-(6,8-dibromo-3-hydroxy-4-oxo-4H-chromen-2-yl)benzoic acid, Casein kinase II subunit alpha | | Authors: | Niefind, K, Bischoff, N, Guerra, B, Golub, A, Issinger, O.-G. | | Deposit date: | 2014-08-12 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | A Note of Caution on the Role of Halogen Bonds for Protein Kinase/Inhibitor Recognition Suggested by High- And Low-Salt CK2 alpha Complex Structures.
Acs Chem.Biol., 10, 2015
|
|
8BYV
 
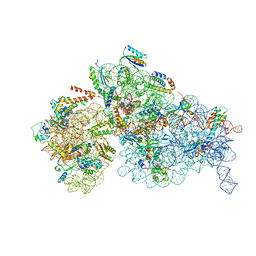 | | Cryo-EM structure of a Staphylococus aureus 30S-RbfA complex | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Bikmullin, A.G, Fatkhullin, B, Stetsenko, A, Guskov, A, Yusupov, M. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Yet Another Similarity between Mitochondrial and Bacterial Ribosomal Small Subunit Biogenesis Obtained by Structural Characterization of RbfA from S. aureus.
Int J Mol Sci, 24, 2023
|
|
1DOG
 
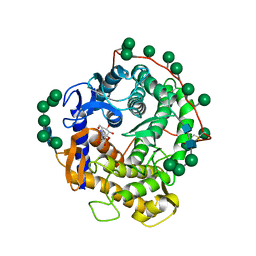 | | REFINED STRUCTURE FOR THE COMPLEX OF 1-DEOXYNOJIRIMYCIN WITH GLUCOAMYLASE FROM (ASPERGILLUS AWAMORI) VAR. X100 TO 2.4 ANGSTROMS RESOLUTION | | Descriptor: | 1-DEOXYNOJIRIMYCIN, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Harris, E, Aleshin, A, Firsov, L, Honzatko, R.B. | | Deposit date: | 1993-01-12 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined structure for the complex of 1-deoxynojirimycin with glucoamylase from Aspergillus awamori var. X100 to 2.4-A resolution.
Biochemistry, 32, 1993
|
|
8BXA
 
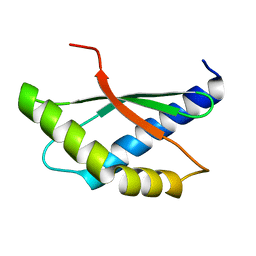 | | Crystal structure of ribosome binding factor A (RbfA) from S. aureus | | Descriptor: | Ribosome-binding factor A | | Authors: | Fatkhullin, B, Bikmullin, A, Gabdulkhakov, A, Khusainov, I, Validov, S, Usachev, K, Yusupov, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Yet Another Similarity between Mitochondrial and Bacterial Ribosomal Small Subunit Biogenesis Obtained by Structural Characterization of RbfA from S. aureus.
Int J Mol Sci, 24, 2023
|
|
6SJ5
 
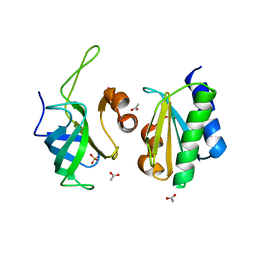 | | Crystal structure of the uL14-RsfS complex from Staphylococcus aureus | | Descriptor: | 50S ribosomal protein L14, ACETIC ACID, PHOSPHATE ION, ... | | Authors: | Fatkhullin, B, Gabdulkhakov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2019-08-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26686549 Å) | | Cite: | Mechanism of ribosome shutdown by RsfS in Staphylococcus aureus revealed by integrative structural biology approach.
Nat Commun, 11, 2020
|
|
6SJ6
 
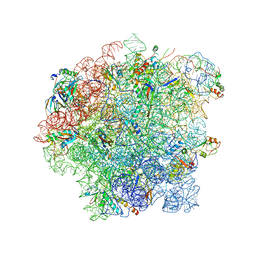 | | Cryo-EM structure of 50S-RsfS complex from Staphylococcus aureus | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Khusainov, I, Pellegrino, S, Yusupova, G, Yusupov, M, Fatkhullin, B. | | Deposit date: | 2019-08-12 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mechanism of ribosome shutdown by RsfS in Staphylococcus aureus revealed by integrative structural biology approach.
Nat Commun, 11, 2020
|
|
1AGM
 
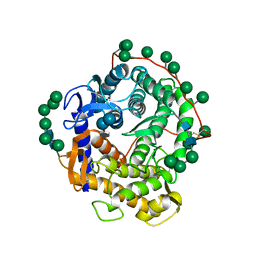 | | Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. x100 to 2.4 angstroms resolution | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Firsov, L.M, Honzatko, R.B. | | Deposit date: | 1994-05-13 | | Release date: | 1994-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. X100 to 2.4-A resolution.
J.Biol.Chem., 269, 1994
|
|
1GAI
 
 | | GLUCOAMYLASE-471 COMPLEXED WITH D-GLUCO-DIHYDROACARBOSE | | Descriptor: | 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Stoffer, B, Firsov, L.M, Svensson, B, Honzatko, R.B. | | Deposit date: | 1996-03-06 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic complexes of glucoamylase with maltooligosaccharide analogs: relationship of stereochemical distortions at the nonreducing end to the catalytic mechanism.
Biochemistry, 35, 1996
|
|
1GAH
 
 | | GLUCOAMYLASE-471 COMPLEXED WITH ACARBOSE | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Stoffer, B, Firsov, L.M, Svensson, B, Honzatko, R.B. | | Deposit date: | 1996-03-06 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic complexes of glucoamylase with maltooligosaccharide analogs: relationship of stereochemical distortions at the nonreducing end to the catalytic mechanism.
Biochemistry, 35, 1996
|
|
3GLY
 
 | | REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 | | Descriptor: | GLUCOAMYLASE-471, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aleshin, A.E, Hoffman, C, Firsov, L.M, Honzatko, R.B. | | Deposit date: | 1994-06-03 | | Release date: | 1994-11-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined crystal structures of glucoamylase from Aspergillus awamori var. X100.
J.Mol.Biol., 238, 1994
|
|
1GLM
 
 | | REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 | | Descriptor: | GLUCOAMYLASE-471, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aleshin, A.E, Hoffman, C, Firsov, L.M, Honzatko, R.B. | | Deposit date: | 1994-04-25 | | Release date: | 1994-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Refined crystal structures of glucoamylase from Aspergillus awamori var. X100.
J.Mol.Biol., 238, 1994
|
|
