5BXZ
 
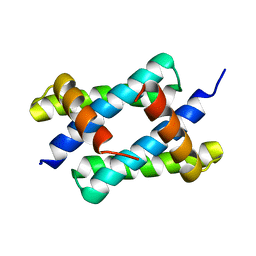 | | H17 Bat Influenza NS1 RNA Binding Domain | | Descriptor: | Non-structural protein 1 | | Authors: | Kerry, P.S, Hale, B.G. | | Deposit date: | 2015-06-09 | | Release date: | 2015-08-19 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel Bat Influenza Virus NS1 Proteins Bind Double-Stranded RNA and Antagonize Host Innate Immunity.
J.Virol., 89, 2015
|
|
5BY1
 
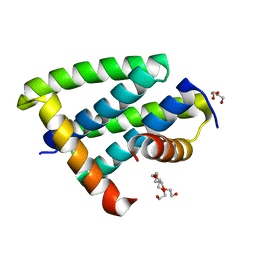 | | H18 Bat Influenza NS1 RNA Binding Domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, Non-structural protein 1, PENTAETHYLENE GLYCOL | | Authors: | Kerry, P.S, Hale, B.G. | | Deposit date: | 2015-06-09 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Novel Bat Influenza Virus NS1 Proteins Bind Double-Stranded RNA and Antagonize Host Innate Immunity.
J.Virol., 89, 2015
|
|
8DIE
 
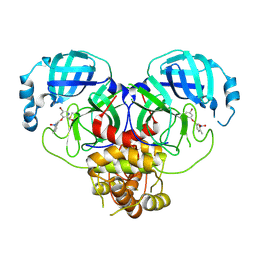 | |
8DIF
 
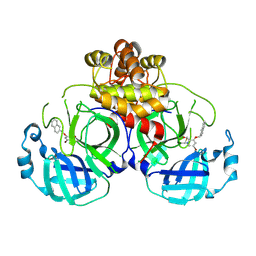 | |
8DIB
 
 | |
8DIG
 
 | |
8DII
 
 | |
8DIH
 
 | | Virtual screening for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors | | Descriptor: | (1P,1'R)-1-(isoquinolin-4-yl)-2',3'-dihydrospiro[imidazolidine-4,1'-indene]-2,5-dione, 3C-like proteinase nsp5 | | Authors: | Singh, I, Shoichet, B.K. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Large library docking for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors.
Protein Sci., 32, 2023
|
|
8DIC
 
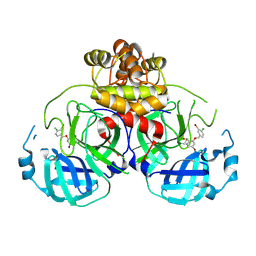 | |
8DID
 
 | |
6E5U
 
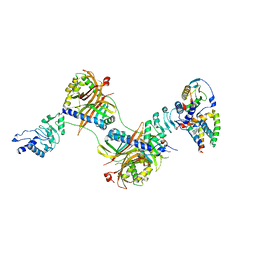 | |
3OA9
 
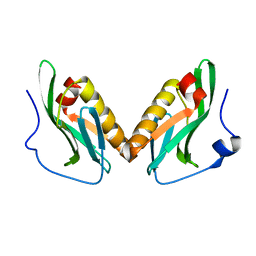 | | Effector domain of influenza A/Duck/Albany/76 NS1 | | Descriptor: | Non-structural protein 1 | | Authors: | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | Deposit date: | 2010-08-05 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
3O9S
 
 | | Effector domain of influenza A/PR/8/34 NS1 | | Descriptor: | Nonstructural protein 1 | | Authors: | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | Deposit date: | 2010-08-04 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
3O9R
 
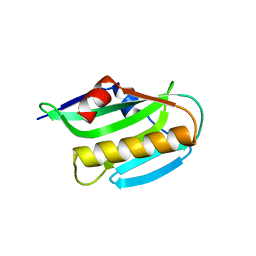 | | Effector domain of NS1 from influenza A/PR/8/34 containing a W187A mutation | | Descriptor: | Nonstructural protein 1 | | Authors: | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | Deposit date: | 2010-08-04 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
3O9Q
 
 | | Effector domain of NS1 from A/PR/8/34 containing a W187A mutation | | Descriptor: | Nonstructural protein 1 | | Authors: | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | Deposit date: | 2010-08-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
3O9T
 
 | | Effector domain from influenza A/PR/8/34 NS1 | | Descriptor: | HEXAETHYLENE GLYCOL, Nonstructural protein 1 | | Authors: | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | Deposit date: | 2010-08-04 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
3O9U
 
 | | Effector domain of influenza A/PR/8/34 NS1 | | Descriptor: | Nonstructural protein 1 | | Authors: | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | Deposit date: | 2010-08-04 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
7GB5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-0da5ad92-2 (Mpro-x10201) | | Descriptor: | 2-(3-chlorophenyl)-N-(pyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.261 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GB3
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with DAV-CRI-3edb475e-6 (Mpro-x10172) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1S)-1-(3-chloro-5-fluorophenyl)ethyl]acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.379 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGZ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-3c65e9ce-2 (Mpro-x12719) | | Descriptor: | 2-(4-acetylpiperazin-1-yl)-N-(4-cyclopropylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBH
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-49816e9b-2 (Mpro-x10334) | | Descriptor: | 2-(3-chlorophenyl)-N-(2,4-dimethylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH0
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-ce760d3f-4 (Mpro-x12723) | | Descriptor: | (4R)-6-chloro-N-(2-oxo-2lambda~5~-isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBT
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-DND-7e92b6ca-2 (Mpro-x10419) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[2-(2-methoxyphenoxy)ethyl]-N-methyl-2-oxo-1,2-dihydroquinoline-4-carboxamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHH
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TOB-UNK-c2aba166-1 (Mpro-x3325) | | Descriptor: | 1-[4-(prop-2-yn-1-yl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GCJ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-590ac91e-19 (Mpro-x10626) | | Descriptor: | (1R,6S,7r)-N-(4-methylpyridin-3-yl)bicyclo[4.1.0]heptane-7-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
