5JW4
 
 | | Structure of MEDI8852 Fab Fragment in Complex with H5 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Neu, U, Collins, P.J, Walker, P.A, Vorlaender, M.K, Ogrodowicz, R.W, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2016-05-11 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes.
Cell, 166, 2016
|
|
7R17
 
 | | Beta Variant SARS-CoV-2 Spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R0Z
 
 | | Dissociated S1 domain of Alpha Variant SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R18
 
 | | Mink Variant SARS-CoV-2 Spike in Closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R12
 
 | | Dissociated S1 domain of Mink Variant SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R1B
 
 | | Mink Variant SARS-CoV-2 Spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R16
 
 | | Beta Variant SARS-CoV-2 Spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R11
 
 | | Dissociated S1 domain of Beta Variant SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R19
 
 | | Mink Variant SARS-CoV-2 Spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R14
 
 | | Alpha Variant SARS-CoV-2 Spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R15
 
 | | Alpha Variant SARS-CoV-2 Spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R13
 
 | | Alpha Variant SARS-CoV-2 Spike in Closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R1A
 
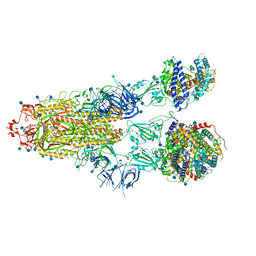 | | Furin Cleaved Alpha Variant SARS-CoV-2 Spike in complex with 3 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R10
 
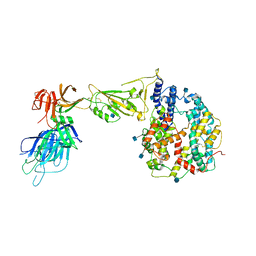 | | Dissociated S1 domain of Alpha Variant SARS-CoV-2 Spike bound to ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
1MXE
 
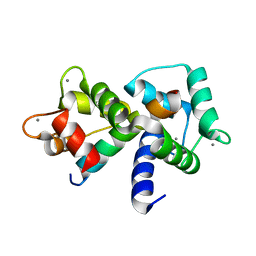 | | Structure of the Complex of Calmodulin with the Target Sequence of CaMKI | | Descriptor: | CALCIUM ION, Calmodulin, Target Sequence of rat Calmodulin-Dependent Protein Kinase I | | Authors: | Clapperton, J.A, Martin, S.R, Smerdon, S.J, Gamblin, S.J, Bayley, P.M. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Complex
of Calmodulin with the Target
Sequence of Calmodulin-Dependent
Protein Kinase I: Studies of the
Kinase Activation Mechanism
Biochemistry, 41, 2002
|
|
1OW3
 
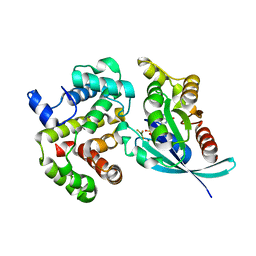 | | Crystal Structure of RhoA.GDP.MgF3-in Complex with RhoGAP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rho-GTPase-activating protein 1, ... | | Authors: | Graham, D.L, Lowe, P.N, Grime, G.W, Marsh, M, Rittinger, K, Smerdon, S.J, Gamblin, S.J, Eccleston, J.F. | | Deposit date: | 2003-03-28 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MgF(3)(-) as a Transition State Analog of Phosphoryl Transfer
Chem.Biol., 9, 2002
|
|
3CKZ
 
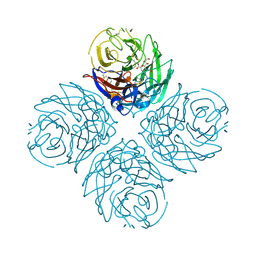 | | N1 Neuraminidase H274Y + Zanamivir | | Descriptor: | CALCIUM ION, Neuraminidase, ZANAMIVIR | | Authors: | Colllins, P, Haire, L.F, Lin, Y.P, Liu, J, Russell, R.J, Walker, P.A, Skehel, J.J, Martin, S.R, Hay, A.J, Gamblin, S.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.
Nature, 453, 2008
|
|
3CL2
 
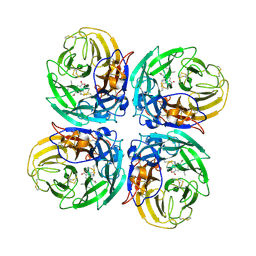 | | N1 Neuraminidase N294S + Oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase | | Authors: | Collins, P, Haire, L.F, Lin, Y.P, Liu, J, Russell, R.J, Walker, P.A, Skehel, J.J, Martin, S.R, Hay, A.J, Gamblin, S.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-20 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.538 Å) | | Cite: | Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.
Nature, 453, 2008
|
|
3CL0
 
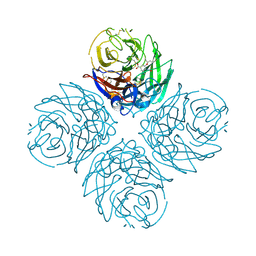 | | N1 Neuraminidase H274Y + oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Collins, P, Haire, L.F, Lin, Y.P, Liu, J, Russell, R.J, Walker, P.A, Skehel, J.J, Martin, S.R, Hay, A.J, Gamblin, S.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-20 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.
Nature, 453, 2008
|
|
1LDN
 
 | | STRUCTURE OF A TERNARY COMPLEX OF AN ALLOSTERIC LACTATE DEHYDROGENASE FROM BACILLUS STEAROTHERMOPHILUS AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, L-LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Wigley, D.B, Gamblin, S.J, Turkenburg, J.P, Dodson, E.J, Piontek, K, Muirhead, H, Holbrook, J.J. | | Deposit date: | 1991-11-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a ternary complex of an allosteric lactate dehydrogenase from Bacillus stearothermophilus at 2.5 A resolution.
J.Mol.Biol., 223, 1992
|
|
3EYM
 
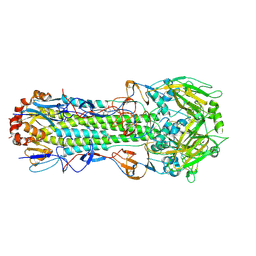 | | Structure of Influenza Haemagglutinin in complex with an inhibitor of membrane fusion | | Descriptor: | 2-tert-butylbenzene-1,4-diol, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Russell, R.J, Kerry, P.S, Stevens, D.A, Steinhauer, D.A, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2008-10-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EYJ
 
 | | Structure of Influenza Haemagglutinin in complex with an inhibitor of membrane fusion | | Descriptor: | Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Russell, R.J, Kerry, P.S, Stevens, D.J, Steinahuer, D.A, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2008-10-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EYK
 
 | | Structure of Influenza Haemagglutinin in complex with an inhibitor of membrane fusion | | Descriptor: | 2-tert-butylbenzene-1,4-diol, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Russell, R.J, Kerry, P.S, Stevens, D.J, Steinhauer, D.A, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2008-10-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3IT5
 
 | | Crystal Structure of the LasA Virulence Factor from Pseudomonas aeruginosa | | Descriptor: | Protease lasA, ZINC ION | | Authors: | Spencer, J, Murphy, L.M, Conners, R, Sessions, R.B, Gamblin, S.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the LasA virulence factor from Pseudomonas aeruginosa: substrate specificity and mechanism of M23 metallopeptidases.
J.Mol.Biol., 396, 2010
|
|
3IIY
 
 | | Crystal structure of Eed in complex with a trimethylated histone H1K26 peptide | | Descriptor: | Histone H1K26 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
