1JVO
 
 | | Azurin dimer, crosslinked via disulfide bridge | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | van Amsterdam, I.M.C, Ubbink, M, Einsle, O, Messerschmidt, A, Merli, A, Cavazzini, D, Rossi, G.L, Canters, G.W. | | Deposit date: | 2001-08-30 | | Release date: | 2002-01-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dramatic modulation of electron transfer in protein complexes by crosslinking
Nat.Struct.Biol., 9, 2002
|
|
2BH4
 
 | | X-ray structure of the M100K variant of ferric cyt c-550 from Paracoccus versutus determined at 100 K. | | Descriptor: | CYTOCHROME C-550, HEME C | | Authors: | Worrall, J.A.R, Van Roon, A.-M.M, Ubbink, M, Canters, G.W. | | Deposit date: | 2005-01-07 | | Release date: | 2005-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Effect of Replacing the Axial Methionine Ligand with a Lysine Residue in Cytochrome C-550 from Paracoccus Versutus Assessed by X-Ray Crystallography and Unfolding.
FEBS J., 272, 2005
|
|
2C6B
 
 | | Solution structure of the C4 zinc-finger domain of HDM2 | | Descriptor: | UBIQUITIN-PROTEIN LIGASE E3 MDM2, ZINC ION | | Authors: | Yu, G.W, Allen, M.D, Andreeva, A, Fersht, A.R, Bycroft, M. | | Deposit date: | 2005-11-08 | | Release date: | 2006-01-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C4 Zinc Finger Domain of Hdm2.
Protein Sci., 15, 2006
|
|
2C3F
 
 | | The structure of a group A streptococcal phage-encoded tail-fibre showing hyaluronan lyase activity. | | Descriptor: | HYALURONIDASE, PHAGE ASSOCIATED, SODIUM ION | | Authors: | Taylor, E.J, Smith, N.L, Linsay, A.-M, Charnock, S.J, Turkenburg, J.P, Dodson, E.J, Davies, G.J, Black, G.W. | | Deposit date: | 2005-10-06 | | Release date: | 2005-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of a Group a Streptococcal Phage-Encoded Virulence Factor Reveals Catalytically Active Triple-Stranded Beta-Helix
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2C1B
 
 | | Structure of cAMP-dependent protein kinase complexed with (4R,2S)-5'-(4-(4-Chlorobenzyloxy)pyrrolidin-2-ylmethanesulfonyl)isoquinoline | | Descriptor: | (4R,2S)-5'-(4-(4-CHLOROBENZYLOXY)PYRROLIDIN-2-YLMETHANESULFONYL)ISOQUINOLINE, CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR | | Authors: | Collins, I, Caldwell, J, Fonseca, T, Donald, A, Bavetsias, V, Hunter, L.J, Garrett, M.D, Rowlands, M.G, Aherne, G.W, Davies, T.G, Berdini, V, Woodhead, S.J, Seavers, L.C.A, Wyatt, P.G, Workman, P, McDonald, E. | | Deposit date: | 2005-09-12 | | Release date: | 2005-11-02 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of isoquinoline-5-sulfonamide inhibitors of protein kinase B.
Bioorg. Med. Chem., 14, 2006
|
|
2C1A
 
 | | Structure of cAMP-dependent protein kinase complexed with Isoquinoline-5-sulfonic acid (2-(2-(4-chlorobenzyloxy)ethylamino) ethyl)amide | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY)ETHYLAMINO)ETHYL)AMIDE | | Authors: | Collins, I, Caldwell, J, Fonseca, T, Donald, A, Bavetsias, V, Hunter, L.J, Garrett, M.D, Rowlands, M.G, Aherne, G.W, Davies, T.G, Berdini, V, Woodhead, S.J, Seavers, L.C.A, Wyatt, P.G, Workman, P, McDonald, E. | | Deposit date: | 2005-09-12 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of isoquinoline-5-sulfonamide inhibitors of protein kinase B.
Bioorg. Med. Chem., 14, 2006
|
|
1KJV
 
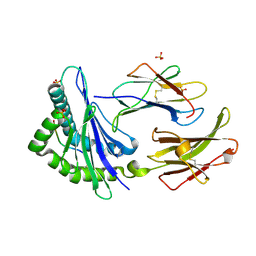 | | TAP-B-associated rat MHC class I molecule | | Descriptor: | Mature alpha chain of major histocompatibility complex class I antigen (HEAVY CHAIN), SULFATE ION, beta-2-microglobulin, ... | | Authors: | Rudolph, M.G, Stevens, J, Speir, J.A, Trowsdale, J, Butcher, G.W, Joly, E, Wilson, I.A. | | Deposit date: | 2001-12-05 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of two rat MHC class Ia (RT1-A) molecules that are associated differentially
with peptide transporter alleles TAP-A and TAP-B.
J.Mol.Biol., 324, 2002
|
|
2BH5
 
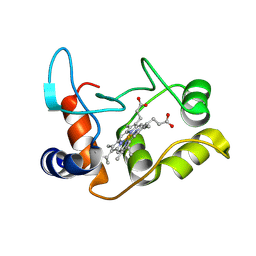 | | X-ray structure of the M100K variant of ferric cyt c-550 from Paracoccus versutus determined at 295 K. | | Descriptor: | CYTOCHROME C-550, HEME C | | Authors: | Worrall, J.A.R, van Roon, A.-M.M, Ubbink, M, Canters, G.W. | | Deposit date: | 2005-01-07 | | Release date: | 2005-05-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Effect of Replacing the Axial Methionine Ligand with a Lysine Residue in Cytochrome C-550 from Paracoccus Versutus Assessed by X-Ray Crystallography and Unfolding.
FEBS J., 272, 2005
|
|
2BGV
 
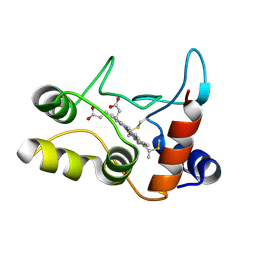 | | X-ray structure of ferric cytochrome c-550 from Paracoccus versutus | | Descriptor: | CYTOCHROME C-550, HEME C | | Authors: | Worrall, J.A.R, Van Roon, A.-M.M, Ubbink, M, Canters, G.W. | | Deposit date: | 2005-01-05 | | Release date: | 2005-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Effect of Replacing the Axial Methionine Ligand with a Lysine Residue in Cytochrome C-550 from Paracoccus Versutus Assessed by X-Ray Crystallography and Unfolding.
FEBS J., 272, 2005
|
|
1KQQ
 
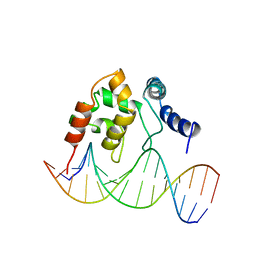 | | Solution Structure of the Dead ringer ARID-DNA Complex | | Descriptor: | 5'-D(*CP*CP*AP*CP*AP*TP*CP*AP*AP*TP*AP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*TP*AP*TP*TP*GP*AP*TP*GP*TP*GP*G)-3', DEAD RINGER PROTEIN | | Authors: | Iwahara, J, Iwahara, M, Daughdrill, G.W, Ford, J, Clubb, R.T. | | Deposit date: | 2002-01-07 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA.
EMBO J., 21, 2002
|
|
1MO7
 
 | | ATPase | | Descriptor: | Sodium/Potassium-transporting ATPase alpha-1 chain | | Authors: | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | Deposit date: | 2002-09-08 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
1MO8
 
 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Sodium/Potassium-Transporting ATPase alpha-1 | | Authors: | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | Deposit date: | 2002-09-08 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
1MZ8
 
 | | CRYSTAL STRUCTURES OF THE NUCLEASE DOMAIN OF COLE7/IM7 IN COMPLEX WITH A PHOSPHATE ION AND A ZINC ION | | Descriptor: | Colicin E7, Colicin E7 immunity protein, PHOSPHATE ION, ... | | Authors: | Sui, M.J, Tsai, L.C, Hsia, K.C, Doudeva, L.G, Ku, W.Y, Han, G.W, Yuan, H.S. | | Deposit date: | 2002-10-07 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal ions and phosphate binding in the H-N-H motif: crystal structures of the nuclease domain of ColE7/Im7 in complex with a phosphate ion and different divalent metal ions
PROTEIN SCI., 11, 2002
|
|
1NPJ
 
 | | Crystal structure of H145A mutant of nitrite reductase from Alcaligenes faecalis | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Wijma, H.J, Boulanger, M.J, Molon, A, Fittipaldi, M, Huber, M, Murphy, M.E, Verbeet, M.P, Canters, G.W. | | Deposit date: | 2003-01-18 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reconstitution of the type-1 active site of the H145G/A variants of nitrite reductase by ligand insertion
Biochemistry, 42, 2003
|
|
1NPN
 
 | | Crystal structure of a copper reconstituted H145A mutant of nitrite reductase from Alcaligenes faecalis | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Wijma, H.J, Boulanger, M.J, Molon, A, Fittipaldi, M, Huber, M, Murphy, M.E, Verbeet, M.P, Canters, G.W. | | Deposit date: | 2003-01-18 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reconstitution of the type-1 active site of the 145G/A variants of Nitrite Reductase by ligand insertion
Biochemistry, 42, 2003
|
|
1OPP
 
 | | PEPTIDE OF HUMAN APOLIPOPROTEIN C-I RESIDUES 1-38, NMR, 28 STRUCTURES | | Descriptor: | APOLIPOPROTEIN C-I | | Authors: | Rozek, A, Buchko, G.W, Kanda, P, Cushley, R.J. | | Deposit date: | 1997-05-08 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational studies of the N-terminal lipid-associating domain of human apolipoprotein C-I by CD and 1H NMR spectroscopy.
Protein Sci., 6, 1997
|
|
1OZI
 
 | | The alternatively spliced PDZ2 domain of PTP-BL | | Descriptor: | protein tyrosine phosphatase | | Authors: | Walma, T, Aelen, J, Oostendorp, M, van den Berk, L, Hendriks, W, Vuister, G.W. | | Deposit date: | 2003-04-09 | | Release date: | 2004-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Closed Binding Pocket and Global Destabilization Modify the Binding Properties of an Alternatively Spliced Form of the Second PDZ Domain of PTP-BL.
Structure, 12, 2004
|
|
1OW3
 
 | | Crystal Structure of RhoA.GDP.MgF3-in Complex with RhoGAP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rho-GTPase-activating protein 1, ... | | Authors: | Graham, D.L, Lowe, P.N, Grime, G.W, Marsh, M, Rittinger, K, Smerdon, S.J, Gamblin, S.J, Eccleston, J.F. | | Deposit date: | 2003-03-28 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MgF(3)(-) as a Transition State Analog of Phosphoryl Transfer
Chem.Biol., 9, 2002
|
|
1P7N
 
 | | Dimeric Rous Sarcoma virus Capsid protein structure with an upstream 25-amino acid residue extension of C-terminal of Gag p10 protein | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27 | | Authors: | Nandhagopal, N, Simpson, A.A, Johnson, M.C, Francisco, A.B, Schatz, G.W, Rossmann, M.G, Vogt, V.M. | | Deposit date: | 2003-05-02 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dimeric rous sarcoma virus capsid protein structure relevant to immature gag assembly
J.Mol.Biol., 335, 2004
|
|
1P9J
 
 | | Solution structure and dynamics of the EGF/TGF-alpha chimera T1E | | Descriptor: | chimera of Epidermal growth factor(EGF) and Transforming growth factor alpha (TGF-alpha) | | Authors: | Wingens, M, Walma, T, Van Ingen, H, Stortelers, C, Van Leeuwen, J.E, Van Zoelen, E.J, Vuister, G.W. | | Deposit date: | 2003-05-12 | | Release date: | 2003-10-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of an Epidermal Growth Factor/Transforming Growth Factor-alpha Chimera with Unique ErbB Binding Specificity.
J.Biol.Chem., 278, 2003
|
|
1PCQ
 
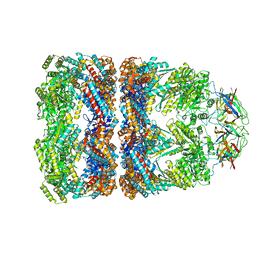 | | Crystal structure of groEL-groES | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Chaudhry, C, Farr, G.W, Todd, M.J, Rye, H.S, Brunger, A.T, Adams, P.D, Horwich, A.L, Sigler, P.B. | | Deposit date: | 2003-05-16 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Role of the gamma-phosphate of ATP in triggering protein folding by GroEL-GroES: function, structure and energetics.
Embo J., 22, 2003
|
|
1PF9
 
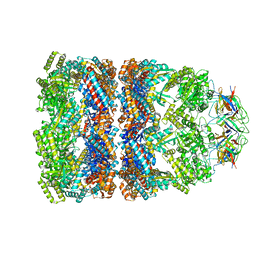 | | GroEL-GroES-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, groEL protein, ... | | Authors: | Chaudhry, C, Farr, G.W, Todd, M.J, Rye, H.S, Brunger, A.T, Adams, P.D, Horwich, A.L, Sigler, P.B. | | Deposit date: | 2003-05-24 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Role of the gamma-phosphate of ATP in triggering protein folding by GroEL-GroES: function, structure and energetics.
Embo J., 22, 2003
|
|
1PD7
 
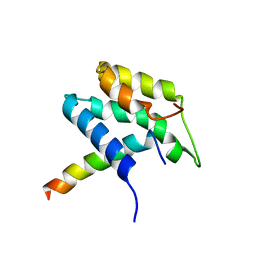 | | Extended SID of Mad1 bound to the PAH2 domain of mSin3B | | Descriptor: | Mad1, Sin3b protein | | Authors: | Van Ingen, H, Lasonder, E, Jansen, J.F, Kaan, A.M, Spronk, C.A, Stunnenberg, H.G, Vuister, G.W. | | Deposit date: | 2003-05-19 | | Release date: | 2004-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Extension of the binding motif of the sin3 interacting domain of the mad family proteins(,).
Biochemistry, 43, 2004
|
|
1PPO
 
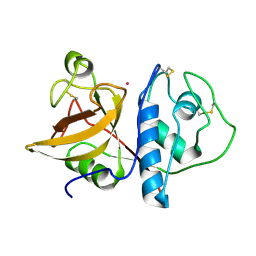 | | DETERMINATION OF THE STRUCTURE OF PAPAYA PROTEASE OMEGA | | Descriptor: | MERCURY (II) ION, PROTEASE OMEGA | | Authors: | Pickersgill, R.W, Rizkallah, P.J, Harris, G.W, Goodenough, P.W. | | Deposit date: | 1991-07-12 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Determination of the Structure of Papaya Protease Omega
Acta Crystallogr.,Sect.B, 47, 1991
|
|
2XF0
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 3-PHENYL-6-(1H-PYRAZOL-4-YL)IMIDAZO[1,2-A]PYRAZINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, McHardy, T, Klair, S, Boxall, K, Fisher, M, Cherry, M, Allen, C.E, Addison, G.J, Ellard, J, Aherne, G.W, Westwood, I.M, van Montfort, R, Garrett, M.D, Reader, J.C, Collins, I. | | Deposit date: | 2010-05-19 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Evaluation of 3,6-Di(Hetero)Aryl Imidazo[1,2-A]Pyrazines as Inhibitors of Checkpoint and Other Kinases.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
