8JOS
 
 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type B crystal | | Descriptor: | Acyltransferase, CHLORIDE ION, TRIETHYLENE GLYCOL | | Authors: | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
8JOR
 
 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type A crystal | | Descriptor: | Acyltransferase, PENTAETHYLENE GLYCOL | | Authors: | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
1ZRN
 
 | | INTERMEDIATE STRUCTURE OF L-2-HALOACID DEHALOGENASE WITH MONOCHLOROACETATE | | Descriptor: | ACETIC ACID, L-2-HALOACID DEHALOGENASE | | Authors: | Li, Y.-F, Hata, Y, Fujii, T, Hisano, T, Nishihara, M, Kurihara, T, Esaki, N. | | Deposit date: | 1998-03-03 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of reaction intermediates of L-2-haloacid dehalogenase and implications for the reaction mechanism.
J.Biol.Chem., 273, 1998
|
|
1ZRM
 
 | | CRYSTAL STRUCTURE OF THE REACTION INTERMEDIATE OF L-2-HALOACID DEHALOGENASE WITH 2-CHLORO-N-BUTYRATE | | Descriptor: | L-2-HALOACID DEHALOGENASE, butanoic acid | | Authors: | Li, Y.-F, Hata, Y, Fujii, T, Hisano, T, Nishihara, M, Kurihara, T, Esaki, N. | | Deposit date: | 1998-03-03 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of reaction intermediates of L-2-haloacid dehalogenase and implications for the reaction mechanism.
J.Biol.Chem., 273, 1998
|
|
8IHL
 
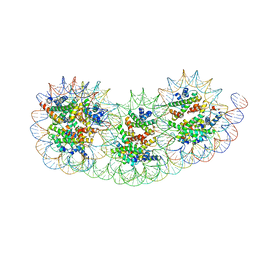 | | Overlapping tri-nucleosome | | Descriptor: | DNA (353-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Fujii, T, Tanaka, H, Maehara, K, Nozawa, K, Takizawa, Y, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (7.64 Å) | | Cite: | Genome-wide mapping and cryo-EM structural analyses of the overlapping tri-nucleosome composed of hexasome-hexasome-octasome moieties.
Commun Biol, 7, 2024
|
|
5ZL1
 
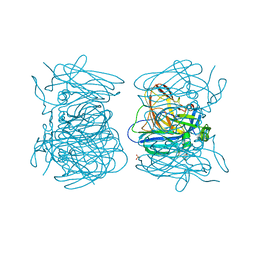 | | Hexameric structure of copper-containing nitrite reductase of an anammox organism KSU-1 | | Descriptor: | CALCIUM ION, COPPER (II) ION, Putative copper-type nitrite reductase, ... | | Authors: | Hira, D, Matsumura, M, Kitamura, R, Fujii, T. | | Deposit date: | 2018-03-26 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hexameric structure of copper-containing nitrite reductase of an anammox organism KSU-1
To Be Published
|
|
1ULK
 
 | | Crystal Structure of Pokeweed Lectin-C | | Descriptor: | lectin-C | | Authors: | Hayashida, M, Fujii, T, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-09-12 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Similarity between protein-protein and protein-carbohydrate interactions, revealed by two crystal structures of lectins from the roots of pokeweed.
J.Mol.Biol., 334, 2003
|
|
1ULM
 
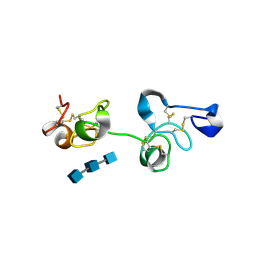 | | Crystal Structure of Pokeweed Lectin-D2 complexed with tri-N-acetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, lectin-D2 | | Authors: | Hayashida, M, Fujii, T, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-09-12 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Similarity between protein-protein and protein-carbohydrate interactions, revealed by two crystal structures of lectins from the roots of pokeweed.
J.Mol.Biol., 334, 2003
|
|
1WPX
 
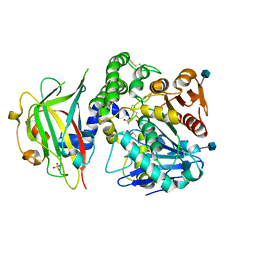 | | Crystal structure of carboxypeptidase Y inhibitor complexed with the cognate proteinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase Y, Carboxypeptidase Y inhibitor, ... | | Authors: | Mima, J, Hayashida, M, Fujii, T, Narita, Y, Hayashi, R, Ueda, M, Hata, Y. | | Deposit date: | 2004-09-14 | | Release date: | 2005-03-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the carboxypeptidase y inhibitor i(c) in complex with the cognate proteinase reveals a novel mode of the proteinase-protein inhibitor interaction
J.Mol.Biol., 346, 2005
|
|
8I2Q
 
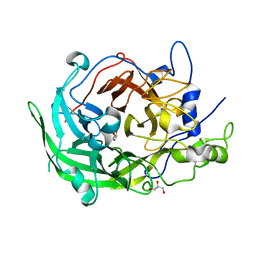 | | Beijerinckia indica beta-fructosyltransferase variant H395R/F473Y | | Descriptor: | Beta-fructosyltransferase, GLYCEROL | | Authors: | Tonozuka, T. | | Deposit date: | 2023-01-15 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Characterization and alteration of product specificity of Beijerinckia indica subsp. indica beta-fructosyltransferase.
Biosci.Biotechnol.Biochem., 87, 2023
|
|
8I2R
 
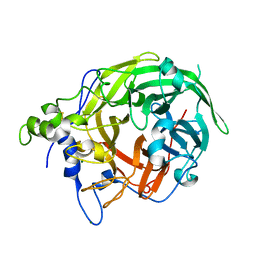 | |
7VCP
 
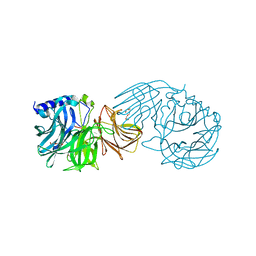 | | Frischella perrara beta-fructofuranosidase in complex with fructose | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Sucrose-6-phosphate hydrolase, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic and structural characterization of beta-fructofuranosidase from the honeybee gut bacterium Frischella perrara.
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7VCO
 
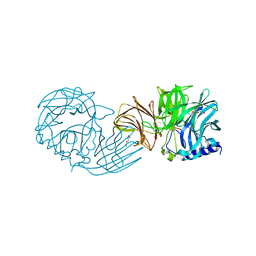 | | Frischella perrara beta-fructofuranosidase | | Descriptor: | DI(HYDROXYETHYL)ETHER, Sucrose-6-phosphate hydrolase, TRIETHYLENE GLYCOL | | Authors: | Tonozuka, T. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzymatic and structural characterization of beta-fructofuranosidase from the honeybee gut bacterium Frischella perrara.
Appl.Microbiol.Biotechnol., 106, 2022
|
|
5WR7
 
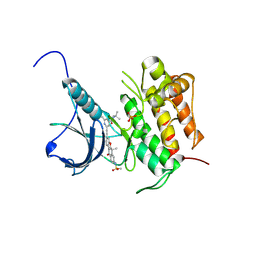 | | Crystal structure of Trk-A complexed with a selective inhibitor CH7057288 | | Descriptor: | High affinity nerve growth factor receptor, N-tert-butyl-2-[2-[6,6-dimethyl-8-(methylsulfonylamino)-11-oxidanylidene-naphtho[2,3-b][1]benzofuran-3-yl]ethynyl]-6-methyl-pyridine-4-carboxamide | | Authors: | Tanaka, H, Blaesse, M, Augustin, M, Goesser, C. | | Deposit date: | 2016-11-30 | | Release date: | 2017-12-06 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Selective TRK Inhibitor CH7057288 against TRK Fusion-Driven Cancer.
Mol. Cancer Ther., 17, 2018
|
|
1ECS
 
 | | THE 1.7 A CRYSTAL STRUCTURE OF A BLEOMYCIN RESISTANCE DETERMINANT ENCODED ON THE TRANSPOSON TN5 | | Descriptor: | BLEOMYCIN RESISTANCE PROTEIN, CALCIUM ION, TETRAETHYLENE GLYCOL | | Authors: | Maruyama, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2000-01-25 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the transposon Tn5-carried bleomycin resistance determinant uncomplexed and complexed with bleomycin.
J.Biol.Chem., 276, 2001
|
|
1EWJ
 
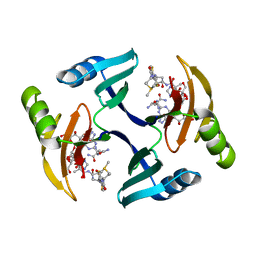 | | CRYSTAL STRUCTURE OF BLEOMYCIN-BINDING PROTEIN COMPLEXED WITH BLEOMYCIN | | Descriptor: | BLEOMYCIN A2, BLEOMYCIN RESISTANCE DETERMINANT | | Authors: | Maruyama, M, Kumagai, T, Matoba, Y, Hata, Y, Sugiyama, M. | | Deposit date: | 2000-04-26 | | Release date: | 2001-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the transposon Tn5-carried bleomycin resistance determinant uncomplexed and complexed with bleomycin.
J.Biol.Chem., 276, 2001
|
|
1KV9
 
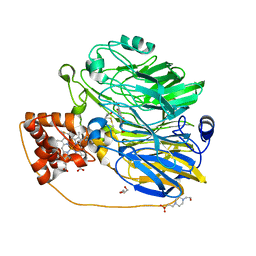 | | Structure at 1.9 A Resolution of a Quinohemoprotein Alcohol Dehydrogenase from Pseudomonas putida HK5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETONE, CALCIUM ION, ... | | Authors: | Chen, Z.-W, Matsushita, K, Yamashita, T, Fujii, T, Toyama, H, Adachi, O, Bellamy, H.D, Mathews, F.S. | | Deposit date: | 2002-01-25 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure at 1.9 A resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas putida HK5.
Structure, 10, 2002
|
|
5B7V
 
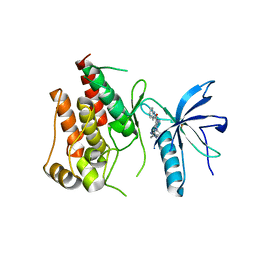 | | Human FGFR1 kinase in complex with CH5183284 | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, [5-amino-1-(2-methyl-1H-benzimidazol-6-yl)-1H-pyrazol-4-yl](1H-indol-2-yl)methanone | | Authors: | Fukami, T.A, Lukacs, C.M, Janson, C. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The fibroblast growth factor receptor genetic status as a potential predictor of the sensitivity to CH5183284/Debio 1347, a novel selective FGFR inhibitor
Mol.Cancer Ther., 13, 2014
|
|
4A62
 
 | |
4A61
 
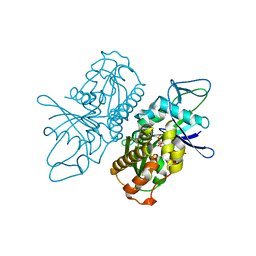 | | ParM from plasmid R1 in complex with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLASMID SEGREGATION PROTEIN PARM | | Authors: | van den Ent, F, Moller-Jensen, J, Gayathri, P, Lowe, J. | | Deposit date: | 2011-10-31 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Bipolar Spindle of Antiparallel Parm Filaments Drives Bacterial Plasmid Segregation.
Science, 338, 2012
|
|
5AZ1
 
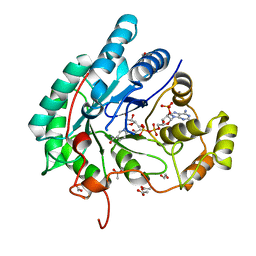 | | Crystal structure of aldo-keto reductase (AKR2E5) complexed with NADPH | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | Deposit date: | 2015-09-15 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5AZ0
 
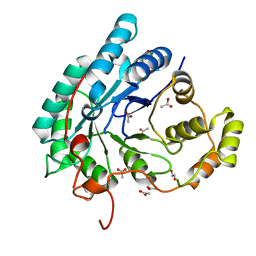 | | Crystal structure of aldo-keto reductase (AKR2E5) of the silkworm, Bombyx mori | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | Deposit date: | 2015-09-15 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori
Biochem.Biophys.Res.Commun., 474, 2016
|
|
1KRM
 
 | |
6M0D
 
 | | Beijerinckia indica beta-fructosyltransferase | | Descriptor: | Levansucrase, MAGNESIUM ION | | Authors: | Tonozuka, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 68 beta-fructosyltransferase from Beijerinckia indica subsp. indica in complex with fructose.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6M0E
 
 | | Beijerinckia indica beta-fructosyltransferase complexed with fructose | | Descriptor: | Levansucrase, MAGNESIUM ION, beta-D-fructofuranose, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 68 beta-fructosyltransferase from Beijerinckia indica subsp. indica in complex with fructose.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
