1VRS
 
 | | Crystal structure of the disulfide-linked complex between the N-terminal and C-terminal domain of the electron transfer catalyst DsbD | | Descriptor: | Thiol:disulfide interchange protein dsbD | | Authors: | Rozhkova, A, Stirnimann, C.U, Frei, P, Grauschopf, U, Brunisholz, R, Gruetter, M.G, Capitani, G, Glockshuber, R. | | Deposit date: | 2005-06-17 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis and kinetics of inter- and intramolecular disulfide exchange in the redox catalyst DsbD
Embo J., 23, 2004
|
|
1P69
 
 | | STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (P417S MUTANT) | | Descriptor: | Coxsackievirus and adenovirus receptor, Fiber protein | | Authors: | Howitt, J, Bewley, M.C, Graziano, V, Flanagan, J.M, Freimuth, P. | | Deposit date: | 2003-04-29 | | Release date: | 2004-05-11 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
J.Biol.Chem., 278, 2003
|
|
1P6A
 
 | | STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (S489Y MUTANT) | | Descriptor: | Coxsackievirus and adenovirus receptor, Fiber protein | | Authors: | Howitt, J, Bewley, M.C, Graziano, V, Flanagan, J.M, Freimuth, P. | | Deposit date: | 2003-04-29 | | Release date: | 2004-05-11 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
J.Biol.Chem., 278, 2003
|
|
4B3S
 
 | | Crystal structure of the 30S ribosome in complex with compound 37 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4-O-benzyl-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B3M
 
 | | Crystal structure of the 30S ribosome in complex with compound 1 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4,6-O-benzylidene-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B3R
 
 | | Crystal structure of the 30S ribosome in complex with compound 30 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-4,6-O-[(1R)-3-phenylpropylidene]-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B3T
 
 | | Crystal structure of the 30S ribosome in complex with compound 39 | | Descriptor: | (2S,3S,4R,5R,6R)-2-(aminomethyl)-5-azanyl-6-[(2R,3S,4R,5S)-5-[(1R,2R,3S,5R,6S)-3,5-bis(azanyl)-2-[(2S,3R,4R,5S,6R)-3-azanyl-5-[(4-chlorophenyl)methoxy]-6-(hydroxymethyl)-4-oxidanyl-oxan-2-yl]oxy-6-oxidanyl-cyclohexyl]oxy-2-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl]oxy-oxane-3,4-diol, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
6HPF
 
 | | Structure of Inactive E165Q mutant of fungal non-CBM carrying GH26 endo-b-mannanase from Yunnania penicillata in complex with alpha-62-61-di-galactosyl-mannotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CHLORIDE ION, ... | | Authors: | von Freiesleben, P, Moroz, O.V, Blagova, E, Wiemann, M, Spodsberg, N, Agger, J.W, Davies, G.J, Wilson, K.S, Stalbrand, H, Meyer, A.S, Krogh, K.B.R.M. | | Deposit date: | 2018-09-20 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure and substrate interactions of an unusual fungal non-CBM carrying GH26 endo-beta-mannanase from Yunnania penicillata.
Sci Rep, 9, 2019
|
|
1NOB
 
 | | KNOB DOMAIN FROM ADENOVIRUS SEROTYPE 12 | | Descriptor: | PROTEIN (FIBER KNOB PROTEIN) | | Authors: | Bewley, M.C, Springer, K, Zhang, Y.B, Freimuth, P, Flanagan, J.M. | | Deposit date: | 1999-05-05 | | Release date: | 1999-11-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR.
Science, 286, 1999
|
|
1KAC
 
 | | KNOB DOMAIN FROM ADENOVIRUS SEROTYPE 12 IN COMPLEX WITH DOMAIN 1 OF ITS CELLULAR RECEPTOR CAR | | Descriptor: | PROTEIN (COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR), PROTEIN (FIBER KNOB PROTEIN) | | Authors: | Bewley, M.C, Springer, K, Zhang, Y.B, Freimuth, P, Flanagan, J.M. | | Deposit date: | 1999-05-05 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR.
Science, 286, 1999
|
|
5MUC
 
 | | Crystal structure of the FimH lectin domain in complex with 1,5-Anhydromannitol | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, Protein FimH | | Authors: | Jakob, R.P, Rabbani, S, Ernst, B, Maier, T. | | Deposit date: | 2017-01-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | KinITC-One Method Supports both Thermodynamic and Kinetic SARs as Exemplified on FimH Antagonists.
Chemistry, 24, 2018
|
|
3F4R
 
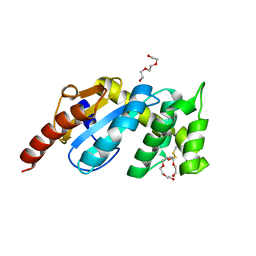 | | Crystal structure of Wolbachia pipientis alpha-DsbA1 | | Descriptor: | PENTAETHYLENE GLYCOL, Putative uncharacterized protein, TRIETHYLENE GLYCOL | | Authors: | Kurz, M, Heras, B, Martin, J.L. | | Deposit date: | 2008-11-02 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of the Oxidoreductase alpha-DsbA1 from Wolbachia pipientis
ANTIOXID.REDOX SIGNAL., 11, 2009
|
|
3BCI
 
 | |
3BD2
 
 | |
3BCK
 
 | |
3F4S
 
 | |
3F4T
 
 | |
5ACG
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
5ACI
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johanson, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
5ACJ
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
1JEW
 
 | |
5ACF
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
4OPB
 
 | | AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Lo Leggio, L, Frandsen, K.H, Davies, G.J, Dupree, P, Walton, P, Henrissat, B, Stringer, M, Tovborg, M, De Maria, L, Johansen, K.S. | | Deposit date: | 2014-02-05 | | Release date: | 2015-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase.
Nat Commun, 6, 2015
|
|
5ACH
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
3TS3
 
 | |
