2NDM
 
 | | NMR solution structure of PawS Derived Peptide 21 (PDP-21) | | Descriptor: | PawS derived peptide 21 | | Authors: | Franke, B, Jayasena, A.S, Fisher, M.F, Swedberg, J.E, Taylor, N.L, Mylne, J.S, Rosengren, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Biopolymers, 106, 2016
|
|
2NDN
 
 | | NMR solution structure of PawS Derived Peptide 20 (PDP-20) | | Descriptor: | PawS1a Derived Peptide 20 | | Authors: | Franke, B, Jayasena, A.S, Fisher, M.F, Swedberg, J.E, Taylor, N.L, Mylne, J.S, Rosengren, K. | | Deposit date: | 2016-07-26 | | Release date: | 2016-08-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Biopolymers, 106, 2016
|
|
2NDL
 
 | | NMR solution structure of PawS Derived Peptide 22 (PDP-22) | | Descriptor: | PawS derived peptide | | Authors: | Franke, B, Jayasena, A.S, Fisher, M.F, Swedberg, J.E, Taylor, N.L, Mylne, J.S, Rosengren, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-12-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Biopolymers, 106, 2016
|
|
5U87
 
 | | NMR structure of the precursor protein PawS1 comprising SFTI-1 and a seed storage albumin | | Descriptor: | Preproalbumin PawS1 | | Authors: | Franke, B, James, A.M, Mobli, M, Colgrave, M.L, Mylne, J.S, Rosengren, K.J. | | Deposit date: | 2016-12-14 | | Release date: | 2017-03-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the precursor protein PawS1 comprising SFTI-1 and a seed storage albumin
to be published
|
|
6YP6
 
 | |
4M3L
 
 | | Crystal Structure of the coiled coil domain of MuRF1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, E3 ubiquitin-protein ligase TRIM63, ... | | Authors: | Mayans, O, Franke, B. | | Deposit date: | 2013-08-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the fold organization and sarcomeric targeting of the muscle atrogin MuRF1.
Open Biol, 4, 2014
|
|
6YGN
 
 | | Titin kinase and its flanking domains | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Titin | | Authors: | Fleming, J.R, Franke, B, Bogomolovas, J, Mayans, O. | | Deposit date: | 2020-03-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Titin kinase ubiquitination aligns autophagy receptors with mechanical signals in the sarcomere.
Embo Rep., 22, 2021
|
|
2NB5
 
 | | NMR solution structure of PawS Derived Peptide 9 (PDP-9) | | Descriptor: | Preproalbumin PawS1 | | Authors: | Armstrong, D.A, Franke, B, Elliott, A.G, Mylne, J.S, Rosengren, K.J. | | Deposit date: | 2016-01-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Natural structural diversity within a conserved cyclic peptide scaffold.
Amino Acids, 49, 2017
|
|
1Y8J
 
 | | Crystal Structure of human NEP complexed with an imidazo[4,5-c]pyridine inhibitor | | Descriptor: | 2-[(1S)-1-BENZYL-2-SULFANYLETHYL]-1H-IMIDAZO[4,5-C]PYRIDIN-5-IUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Sahli, S, Frank, B, Schweizer, W.B, Diederich, F, Blum-Kaelin, D, Aebi, J.D, Bohm, H.J, Oefner, C, Dale, G.E. | | Deposit date: | 2004-12-13 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Second-Generation Inhibitors for the Metalloprotease Neprilysin Based on Bicyclic Heteroaromatic Scaffolds: Synthesis, Biological Activity, and X-ray Crystal Structure Analysis
HELV.CHIM.ACTA, 88, 2005
|
|
5JDE
 
 | |
5JDD
 
 | |
5JDJ
 
 | | Crystal structure of domain I10 from titin in space group P212121 | | Descriptor: | CALCIUM ION, Titin | | Authors: | Williams, R, Bogomolovas, J, Labiet, S, Mayans, O. | | Deposit date: | 2016-04-16 | | Release date: | 2016-08-17 | | Last modified: | 2016-10-12 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Exploration of pathomechanisms triggered by a single-nucleotide polymorphism in titin's I-band: the cardiomyopathy-linked mutation T2580I.
Open Biology, 6, 2016
|
|
3UTO
 
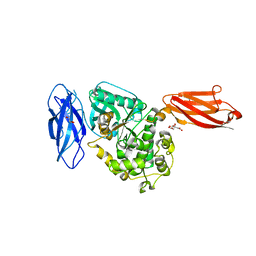 | | Twitchin kinase region from C.elegans (Fn31-NL-kin-CRD-Ig26) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Castelmur, E, Barbieri, S, Mayans, O. | | Deposit date: | 2011-11-26 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of an N-terminal inhibitory extension as the primary mechanosensory regulator of twitchin kinase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5JOE
 
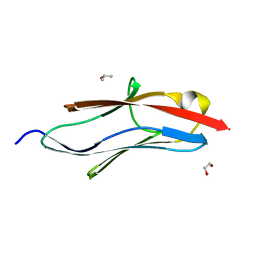 | | Crystal structure of I81 from titin | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Titin | | Authors: | Fleming, J, Zhou, T, Bogomolovas, J, Labeit, S, Mayans, O. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARP interacts with titin at a unique helical N2A sequence and at the domain Ig81 to form a structured complex.
Febs Lett., 590, 2016
|
|
2NB6
 
 | |
4L02
 
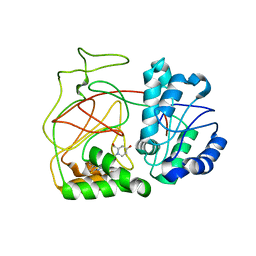 | | Crystal Structure of SphK1 with inhibitor | | Descriptor: | (2R,4S)-1-[2-(4-{[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]amino}phenyl)ethyl]-2-(hydroxymethyl)piperidin-4-ol, Sphingosine kinase 1 | | Authors: | Min, X, Walker, N, Wang, Z. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure guided design of a series of sphingosine kinase (SphK) inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7L51
 
 | |
7L55
 
 | |
7L54
 
 | |
7L53
 
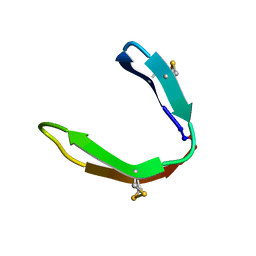 | |
