2CSA
 
 | | Structure of the M3 Muscarinic Acetylcholine Receptor Basolateral Sorting Signal | | Descriptor: | Muscarinic acetylcholine receptor M3 | | Authors: | Iverson, H.A, Fox, D, Nadler, L.S, Klevit, R.E, Nathanson, N.M. | | Deposit date: | 2005-05-21 | | Release date: | 2005-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Identification and structural determination of the M3 muscarinic acetylcholine receptor basolateral sorting signal.
J.Biol.Chem., 280, 2005
|
|
4F2G
 
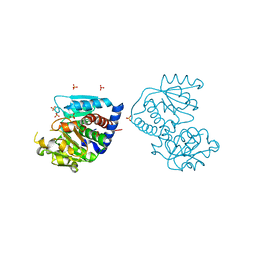 | | The Crystal Structure of Ornithine carbamoyltransferase from Burkholderia thailandensis E264 | | Descriptor: | 1,2-ETHANEDIOL, Ornithine carbamoyltransferase 1, PHOSPHATE ION | | Authors: | Craig, T.K, Fox, D, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
4MYQ
 
 | |
9DIR
 
 | | Cryo-EM structure of the heme/hemoglobin transporter ChuA, in complex with de novo designed binder G7 | | Descriptor: | ChuA binding protein G7, Outer membrane heme/hemoglobin receptor | | Authors: | Fox, D, Venugopal, H, Lupton, C.J, Spicer, B.A, Grinter, R. | | Deposit date: | 2024-09-05 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Inhibiting heme piracy by pathogenic Escherichia coli using de novo-designed proteins
Nat Commun, 2025
|
|
9DIV
 
 | |
9DIS
 
 | | Cryo-EM structure of the heme/hemoglobin transporter ChuA, in complex with de novo designed binder H3 | | Descriptor: | ChuA Binder H3, Outer membrane heme/hemoglobin receptor | | Authors: | Fox, D, Venugopal, H, Lupton, C.J, Spicer, B.A, Grinter, R. | | Deposit date: | 2024-09-05 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Inhibiting heme piracy by pathogenic Escherichia coli using de novo-designed proteins
Nat Commun, 2025
|
|
9DHE
 
 | |
3C5R
 
 | |
8VYL
 
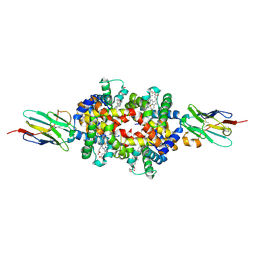 | | The structure of Human Hemoglobin in Complex with Nanobody BtNbE11 | | Descriptor: | ACETYL GROUP, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Grinter, R, Binks, S, Fox, D. | | Deposit date: | 2024-02-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The structure of a haemoglobin-nanobody complex reveals human beta-subunit-specific interactions.
Febs Lett., 598, 2024
|
|
6C99
 
 | | Crystal structure of FcRn bound to UCB-303 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, CITRIC ACID, ... | | Authors: | Fox III, D, Abendroth, J, Porter, J, Deboves, H. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
6C97
 
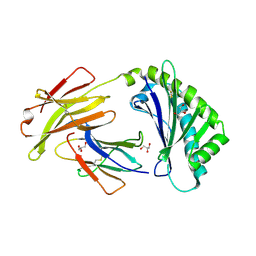 | | Crystal structure of FcRn at pH3 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, IgG receptor FcRn large subunit p51 | | Authors: | Fox III, D, Fairman, J.W. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
6C98
 
 | | Crystal structure of FcRn bound to UCB-84 | | Descriptor: | 1-[7-(3-fluorophenyl)-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-6-yl]ethan-1-one, Beta-2-microglobulin, CYSTEINE, ... | | Authors: | Fox III, D, Lukacs, C.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
4DXL
 
 | |
4WTD
 
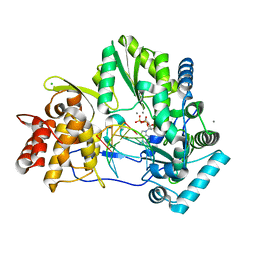 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS AND DELTA8 BETA HAIRPIN LOOP DELETION IN COMPLEX WITH ADP, MN2+ AND SYMMETRICAL PRIMER TEMPLATE 5'-AUAAAUUU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Edwards, T.E, Fox III, D, Appleby, T.C. | | Deposit date: | 2014-10-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTM
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-UAGG, RNA PRIMER 5'-PCC, MN2+, AND UDP | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, RNA PRIMER CC, ... | | Authors: | Edwards, T.E, Appleby, T.C, Fox III, D. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4DZ2
 
 | |
3O38
 
 | |
3OC6
 
 | |
3OI9
 
 | |
3P2Y
 
 | |
3OME
 
 | |
3OKS
 
 | |
4DUT
 
 | |
4DZ4
 
 | |
4E4T
 
 | |
