1MMD
 
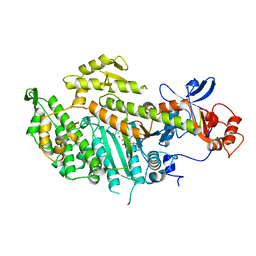 | | TRUNCATED HEAD OF MYOSIN FROM DICTYOSTELIUM DISCOIDEUM COMPLEXED WITH MGADP-BEF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Fisher, A.J, Holden, H.M, Rayment, I. | | Deposit date: | 1995-03-21 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4-.
Biochemistry, 34, 1995
|
|
1LUC
 
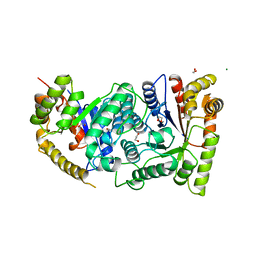 | | BACTERIAL LUCIFERASE | | Descriptor: | 1,2-ETHANEDIOL, BACTERIAL LUCIFERASE, MAGNESIUM ION | | Authors: | Fisher, A.J, Rayment, I. | | Deposit date: | 1996-05-10 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5-A resolution crystal structure of bacterial luciferase in low salt conditions.
J.Biol.Chem., 271, 1996
|
|
1P35
 
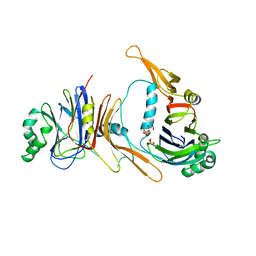 | | CRYSTAL STRUCTURE OF BACULOVIRUS P35 | | Descriptor: | 1,2-ETHANEDIOL, P35, PHOSPHATE ION | | Authors: | Fisher, A.J, Delacruz, W.P, Zoog, S.J, Schneider, C.L, Friesen, P.D. | | Deposit date: | 1998-09-18 | | Release date: | 1999-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of baculovirus P35: role of a novel reactive site loop in apoptotic caspase inhibition.
EMBO J., 18, 1999
|
|
1BRL
 
 | |
9DP5
 
 | |
8E4X
 
 | |
6BGB
 
 | |
7TNI
 
 | |
7KL5
 
 | | Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues: Phe4246 to Val4271 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calmodulin-1, ... | | Authors: | Fisher, A.J, Ames, J.B, Yu, Q. | | Deposit date: | 2020-10-29 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues Phe4246-Val4271 Reveals a Fifth Calcium Binding Site.
Biochemistry, 60, 2021
|
|
6VLB
 
 | |
6VLC
 
 | |
9D5K
 
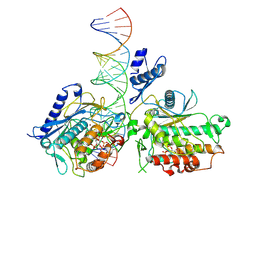 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2-RD) bound to dsRNA containing an expanded cytidine analog at the -1 position of the guide strand | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Isoform 4 of Double-stranded RNA-specific editase 1, MAGNESIUM ION, ... | | Authors: | Fisher, A.J, Cheng, J, Manjunath, A, Campbell, K. | | Deposit date: | 2024-08-13 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Nucleoside Analogs in ADAR Guide Strands Enable Editing at 5'-G A Sites.
Biomolecules, 14, 2024
|
|
9D5J
 
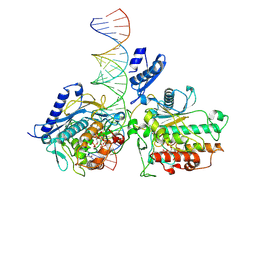 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2-RD) bound to dsRNA containing deoxyinosine at the -1 position of the guide strand | | Descriptor: | 1,2-ETHANEDIOL, INOSITOL HEXAKISPHOSPHATE, Isoform 4 of Double-stranded RNA-specific editase 1, ... | | Authors: | Fisher, A.J, Cheng, J, Manjunath, A, Campbell, K. | | Deposit date: | 2024-08-13 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleoside Analogs in ADAR Guide Strands Enable Editing at 5'-G A Sites.
Biomolecules, 14, 2024
|
|
4IME
 
 | |
4IMC
 
 | |
4IMG
 
 | |
4IMD
 
 | |
4IMF
 
 | |
5DJI
 
 | | Structure of M. tuberculosis CysQ, a PAP phosphatase with AMP, PO4, and 2Mg bound | | Descriptor: | 3'-phosphoadenosine 5'-phosphate phosphatase, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fisher, A.J, Erickson, A.I. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Crystal Structures of Mycobacterium tuberculosis CysQ, with Substrate and Products Bound.
Biochemistry, 54, 2015
|
|
5DJJ
 
 | | Structure of M. tuberculosis CysQ, a PAP phosphatase with PO4 and 2Mg bound | | Descriptor: | 3'-phosphoadenosine 5'-phosphate phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fisher, A.J, Erickson, A.I. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal Structures of Mycobacterium tuberculosis CysQ, with Substrate and Products Bound.
Biochemistry, 54, 2015
|
|
5DJG
 
 | | Structure of M. tuberculosis CysQ, a PAP phosphatase with PAP, Mg, and Li bound | | Descriptor: | 3'-phosphoadenosine 5'-phosphate phosphatase, ADENOSINE-3'-5'-DIPHOSPHATE, LITHIUM ION, ... | | Authors: | Fisher, A.J, Erickson, A.I. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystal Structures of Mycobacterium tuberculosis CysQ, with Substrate and Products Bound.
Biochemistry, 54, 2015
|
|
5DJH
 
 | | Structure of M. tuberculosis CysQ, a PAP phosphatase with AMP, PO4, and 3Mg bound | | Descriptor: | 3'-phosphoadenosine 5'-phosphate phosphatase, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fisher, A.J, Erickson, A.I. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Crystal Structures of Mycobacterium tuberculosis CysQ, with Substrate and Products Bound.
Biochemistry, 54, 2015
|
|
5DJK
 
 | | Structure of M. tuberculosis CysQ, a PAP phosphatase with PO4 and 2Ca bound | | Descriptor: | 3'-phosphoadenosine 5'-phosphate phosphatase, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Fisher, A.J, Erickson, A.I. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal Structures of Mycobacterium tuberculosis CysQ, with Substrate and Products Bound.
Biochemistry, 54, 2015
|
|
4R37
 
 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 with UDP-GlcNAc | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Fisher, A.J, Chen, X, Ngo, A. | | Deposit date: | 2014-08-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R83
 
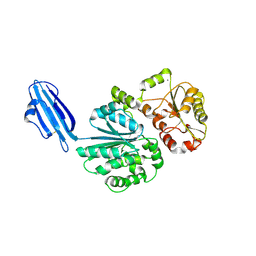 | | Crystal structure of Sialyltransferase from Photobacterium damsela | | Descriptor: | CALCIUM ION, Sialyltransferase 0160 | | Authors: | Fisher, A.J, Chen, X, Li, Y, Huynh, N. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
