4IA6
 
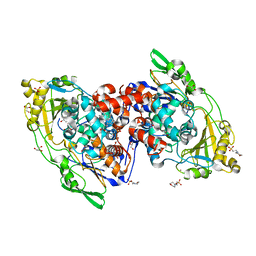 | | Hydratase from lactobacillus acidophilus in a ligand bound form (LA LAH) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Khoshnevis, S, Neumann, P, Ficner, R. | | Deposit date: | 2012-12-06 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of a fatty acid double-bond hydratase from Lactobacillus acidophilus
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5LZB
 
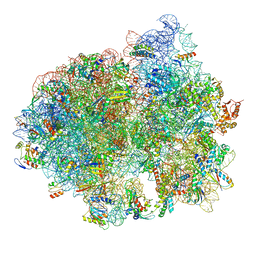 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the initial binding state (IB) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
4H69
 
 | |
4FEE
 
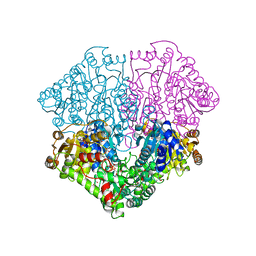 | | High-resolution structure of pyruvate oxidase in complex with reaction intermediate 2-hydroxyethyl-thiamin diphosphate carbanion-enamine, crystal B | | Descriptor: | 2-[(2E)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-HYDROXYETHYLIDENE)-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Meyer, D, Neumann, P, Koers, E, Sjuts, H, Luedtke, S, Sheldrick, G.M, Ficner, R, Tittmann, K. | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Unexpected tautomeric equilibria of the carbanion-enamine intermediate in pyruvate oxidase highlight unrecognized chemical versatility of thiamin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5MED
 
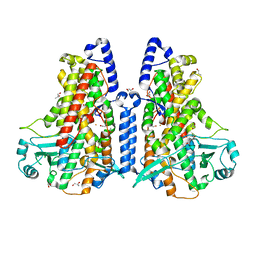 | | Cyanothece lipoxygenase 2 (CspLOX2) | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, 1-BUTANOL, ... | | Authors: | Newie, J, Neumann, P, Werner, M, Mata, R.A, Ficner, R, Feussner, I. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.
Sci Rep, 7, 2017
|
|
4FEG
 
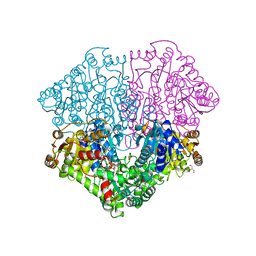 | | High-resolution structure of pyruvate oxidase in complex with reaction intermediate 2-hydroxyethyl-thiamin diphosphate carbanion-enamine, crystal A | | Descriptor: | 2-[(2E)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-HYDROXYETHYLIDENE)-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Meyer, D, Neumann, P, Koers, E, Sjuts, H, Luedtke, S, Sheldrick, G.M, Ficner, R, Tittmann, K. | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Unexpected tautomeric equilibria of the carbanion-enamine intermediate in pyruvate oxidase highlight unrecognized chemical versatility of thiamin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HZK
 
 | |
4IA5
 
 | | Hydratase from Lactobacillus acidophilus - SeMet derivative (apo LAH) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Khoshnevis, S, Neumann, P, Ficner, R. | | Deposit date: | 2012-12-06 | | Release date: | 2013-03-27 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure analysis of a fatty acid double-bond hydratase from Lactobacillus acidophilus
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3NS5
 
 | |
3JQY
 
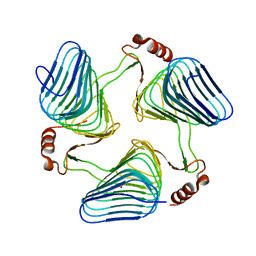 | | Crystal Structure of the polySia specific acetyltransferase NeuO | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Polysialic acid O-acetyltransferase | | Authors: | Schulz, E.-C, Bergfeld, A, Muehlenhoff, M, Ficner, R. | | Deposit date: | 2009-09-08 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure analysis of the polysialic acid specific O-acetyltransferase NeuO
PLoS ONE, 6, 2011
|
|
3GJX
 
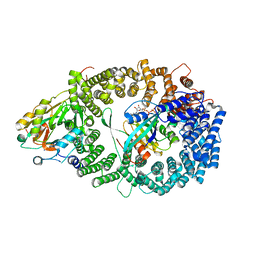 | | Crystal Structure of the Nuclear Export Complex CRM1-Snurportin1-RanGTP | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Monecke, T, Guettler, T, Neumann, P, Dickmanns, A, Goerlich, D, Ficner, R. | | Deposit date: | 2009-03-09 | | Release date: | 2009-05-26 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Nuclear Export Receptor CRM1 in Complex with Snurportin1 and RanGTP.
Science, 2009
|
|
3I4U
 
 | |
3G8V
 
 | |
3G91
 
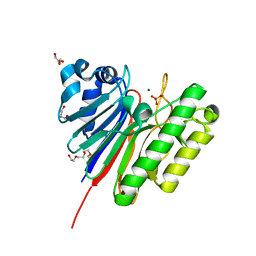 | | 1.2 Angstrom structure of the exonuclease III homologue Mth0212 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-12 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3NC0
 
 | | Crystal structure of the HIV-1 Rev NES-CRM1-RanGTP nuclear export complex (crystal II) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Exportin-1, GLYCEROL, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NBY
 
 | | Crystal structure of the PKI NES-CRM1-RanGTP nuclear export complex | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NBZ
 
 | | Crystal structure of the HIV-1 Rev NES-CRM1-RanGTP nuclear export complex (crystal I) | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NS6
 
 | |
1YY3
 
 | |
3NC1
 
 | | Crystal structure of the CRM1-RanGTP complex | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1Y4J
 
 | | Crystal structure of the paralogue of the human formylglycine generating enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dickmanns, A, Rudolph, M.G, Ficner, R. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Crystal Structure of Human pFGE, the Paralog of the C{alpha}-formylglycine-generating Enzyme
J.Biol.Chem., 280, 2005
|
|
3OGZ
 
 | | Protein structure of USP from L. major in Apo-form | | Descriptor: | GLYCEROL, UDP-sugar pyrophosphorylase | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
1Y1E
 
 | | human formylglycine generating enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-alpha-formyglycine-generating enzyme, CALCIUM ION | | Authors: | Rudolph, M.G, Dickmanns, A, Ficner, R. | | Deposit date: | 2004-11-18 | | Release date: | 2005-05-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular basis for multiple sulfatase deficiency and mechanism for formylglycine generation of the human formylglycine-generating enzyme.
Cell(Cambridge,Mass.), 121, 2005
|
|
1Y1I
 
 | | hyuman formylglycine generating enzyme, reduced form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-alpha-formyglycine-generating enzyme, CALCIUM ION | | Authors: | Rudolph, M.G, Dickmanns, A, Ficner, R. | | Deposit date: | 2004-11-18 | | Release date: | 2005-05-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Molecular basis for multiple sulfatase deficiency and mechanism for formylglycine generation of the human formylglycine-generating enzyme.
Cell(Cambridge,Mass.), 121, 2005
|
|
1Y1G
 
 | | Human formylglycine generating enzyme, double sulfonic acid form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-alpha-formyglycine-generating enzyme, CALCIUM ION | | Authors: | Rudolph, M.G, Dickmanns, A, Ficner, R. | | Deposit date: | 2004-11-18 | | Release date: | 2005-05-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Molecular basis for multiple sulfatase deficiency and mechanism for formylglycine generation of the human formylglycine-generating enzyme.
Cell(Cambridge,Mass.), 121, 2005
|
|
