2JGP
 
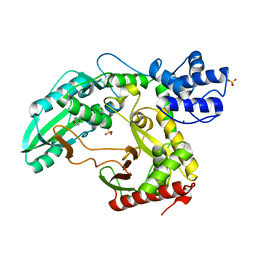 | | Structure of the TycC5-6 PCP-C bidomain of the tyrocidine synthetase TycC | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, SODIUM ION, SULFATE ION, ... | | Authors: | Samel, S.A, Schoenafinger, G, Knappe, T.A, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2007-02-13 | | Release date: | 2007-10-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Insights Into a Peptide Bond-Forming Bidomain from a Nonribosomal Peptide Synthetase.
Structure, 15, 2007
|
|
2JJA
 
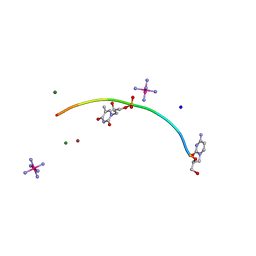 | | Crystal structure of GNA with synthetic copper base pair | | Descriptor: | COBALT HEXAMMINE(III), COPPER (II) ION, GNA, ... | | Authors: | Schlegel, M.K, Essen, L.-O, Meggers, E. | | Deposit date: | 2008-03-28 | | Release date: | 2008-07-22 | | Last modified: | 2022-06-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Duplex Structure of a Minimal Nucleic Acid.
J.Am.Chem.Soc., 130, 2008
|
|
1E0R
 
 | | Beta-apical domain of thermosome | | Descriptor: | THERMOSOME | | Authors: | Bosch, G, Baumeister, W, Essen, L.-O. | | Deposit date: | 2000-04-06 | | Release date: | 2000-08-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Beta-Apical Domain from Thermosome Reveals Structural Plasticity in Protrusion Region
J.Mol.Biol., 301, 2000
|
|
7ZHF
 
 | |
7ZHK
 
 | |
2XC6
 
 | |
2WNA
 
 | |
2VTB
 
 | | Structure of cryptochrome 3 - DNA complex | | Descriptor: | 5'-D(*DT*DT*DT*DT*DTP)-3', 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, ACETATE ION, ... | | Authors: | Pokorny, R, Klar, T, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2008-05-13 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Recognition and Repair of Uv Lesions in Loop Structures of Duplex DNA by Dash-Type Cryptochrome.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3ZXS
 
 | | Cryptochrome B from Rhodobacter sphaeroides | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, CRYPTOCHROME B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Geisselbrecht, Y, Fruhwirth, S, Pierik, A.J, Klug, G, Essen, L.-O. | | Deposit date: | 2011-08-15 | | Release date: | 2012-02-15 | | Last modified: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cryb from Rhodobacter Sphaeroides: A Unique Class of Cryptochromes with New Cofactors.
Embo Rep., 13, 2012
|
|
4AHW
 
 | |
4AI0
 
 | |
4AI3
 
 | |
4AHY
 
 | | Flo5A cocrystallized with 3 mM GdAc3 | | Descriptor: | CHLORIDE ION, FLOCCULATION PROTEIN FLO5, GADOLINIUM ATOM | | Authors: | Veelders, M, Essen, L.-O. | | Deposit date: | 2012-02-07 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Complex Gadolinium-Oxo Clusters Formed Along Concave Protein Surfaces.
Chembiochem, 13, 2012
|
|
4AI1
 
 | |
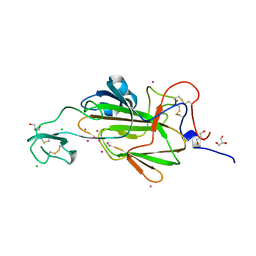
4AHZ
 
 | |
4AI2
 
 | |
4ASL
 
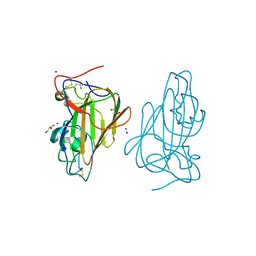 | | Structure of Epa1A in complex with the T-antigen (Gal-b1-3- GalNAc) | | Descriptor: | CALCIUM ION, EPA1P, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Diderrich, R, Veelders, M.S, Eulenburg, G, Kalugin, V, Brueckner, S, Keller, P, Rupp, S, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2012-05-02 | | Release date: | 2012-10-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural Basis for Promiscuity and Specificity During Candida Glabrata Invasion of Host Epithelia.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AS0
 
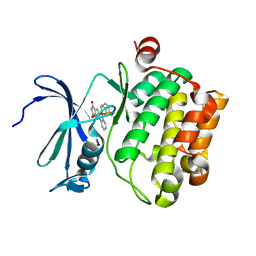 | | Cyclometalated Phthalimides as Protein Kinase Inhibitors | | Descriptor: | PHTALIMIDE-RUTHENIUM COMPLEX, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Blanck, S, Geisselbrecht, Y, Middel, S, Mietke, T, Harms, K, Essen, L.-O, Meggers, E. | | Deposit date: | 2012-04-27 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bioactive Cyclometalated Phthalimides: Design, Synthesis and Kinase Inhibition.
Dalton Trans, 41, 2012
|
|
4BWI
 
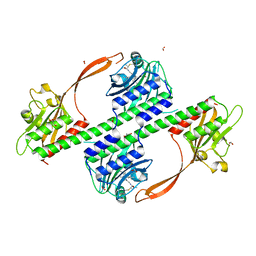 | | Structure of the phytochrome Cph2 from Synechocystis sp. PCC6803 | | Descriptor: | FORMIC ACID, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Anders, K, Angerer, V, Widany, G.D, Mroginski, M.A, von Stetten, D, Essen, L.-O. | | Deposit date: | 2013-07-03 | | Release date: | 2013-10-30 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Cyanobacterial Phytochrome 2 Photosensor Implies a Tryptophan Switch for Phytochrome Signaling.
J.Biol.Chem., 288, 2013
|
|
4CP1
 
 | | Crystal Structure of Epithelial Adhesin 9 A domain (Epa9A) from Candida glabrata in complex with Galb1-3GlcNAc | | Descriptor: | CALCIUM ION, CHLORIDE ION, EPITHELIAL ADHESIN 9, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional reprogramming of Candida glabrata epithelial adhesins: the role of conserved and variable structural motifs in ligand binding
To Be Published
|
|
4COW
 
 | | Crystal Structure of Epithelial Adhesin 6 A domain (Epa6A) from Candida glabrata in complex with the T-antigen (Galb1-3GalNAc) | | Descriptor: | ACETATE ION, CALCIUM ION, EPITHELIAL ADHESIN 6, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
4COV
 
 | | Crystal Structure of Epithelial Adhesin 6 A domain (Epa6A) from Candida glabrata in complex with Gala1-3Gal | | Descriptor: | ACETATE ION, CALCIUM ION, EPITHELIAL ADHESIN 6, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
4COZ
 
 | | Crystal Structure of Epithelial Adhesin 6 A domain (Epa6A) from Candida glabrata in complex with Galb1-3GlcNAc | | Descriptor: | ACETATE ION, CALCIUM ION, EPITHELIAL ADHESIN 6, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
4COY
 
 | | Crystal Structure of Epithelial Adhesin 6 A domain (Epa6A) from Candida glabrata in complex with Galb1-4GlcNAc | | Descriptor: | ACETATE ION, CALCIUM ION, EPITHELIAL ADHESIN 6, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
4D3W
 
 | | Crystal Structure of Epithelial Adhesin 1 A domain (Epa1A) from Candida glabrata in complex with the T-antigen (Galb1-3GalNAc) | | Descriptor: | CALCIUM ION, EPITHELIAL ADHESIN 1, GLYCEROL, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-10-24 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
