8GCL
 
 | | Cryo-EM structure of hAQP2 in DDM | | Descriptor: | Aquaporin-2 | | Authors: | Kamegawa, A, Suzuki, S, Nishikawa, K, Numoto, N, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-03-02 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural analysis of the water channel AQP2 by single-particle cryo-EM.
J.Struct.Biol., 215, 2023
|
|
5JR0
 
 | |
8JT7
 
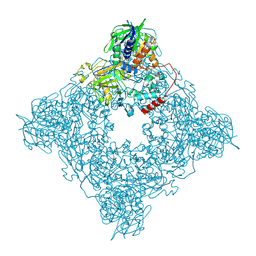 | | Structure of arginine oxidase from Pseudomonas sp. TRU 7192 | | Descriptor: | Amine oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yamaguchi, H, Numoto, N, Suzuki, H, Nishikawa, K, Kamegawa, A, Takahashi, K, Sugiki, M, Fujiyoshi, Y. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Structural basis of arginine oxidase from Pseudomonas sp. TRU 7192
To Be Published
|
|
2D57
 
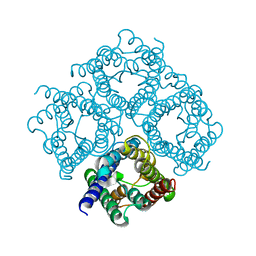 | | Double layered 2D crystal structure of AQUAPORIN-4 (AQP4M23) at 3.2 a resolution by electron crystallography | | Descriptor: | Aquaporin-4 | | Authors: | Hiroaki, Y, Tani, K, Kamegawa, A, Gyobu, N, Nishikawa, K, Suzuki, H, Walz, T, Sasaki, S, Mitsuoka, K, Kimura, K, Mizoguchi, A, Fujiyoshi, Y. | | Deposit date: | 2005-10-29 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.2 Å) | | Cite: | Implications of the Aquaporin-4 Structure on Array Formation and Cell Adhesion
J.Mol.Biol., 355, 2005
|
|
6ACF
 
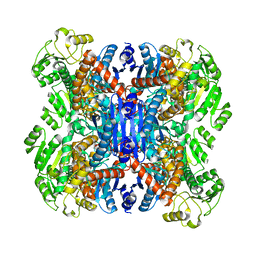 | | structure of leucine dehydrogenase from Geobacillus stearothermophilus by cryo-EM | | Descriptor: | Leucine dehydrogenase | | Authors: | Yamaguchi, H, Kamegawa, A, Nakata, K, Kashiwagi, T, Mizukoshi, T, Fujiyoshi, Y, Tani, K. | | Deposit date: | 2018-07-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into thermostabilization of leucine dehydrogenase from its atomic structure by cryo-electron microscopy
J. Struct. Biol., 205, 2019
|
|
6ACH
 
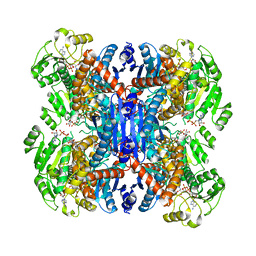 | | Structure of NAD+-bound leucine dehydrogenase from Geobacillus stearothermophilus by cryo-EM | | Descriptor: | Leucine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamaguchi, H, Kamegawa, A, Nakata, K, Kashiwagi, T, Mizukoshi, T, Fujiyoshi, Y, Tani, K. | | Deposit date: | 2018-07-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into thermostabilization of leucine dehydrogenase from its atomic structure by cryo-electron microscopy
J. Struct. Biol., 205, 2019
|
|
3IYZ
 
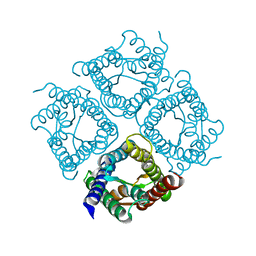 | | Structure of Aquaporin-4 S180D mutant at 10.0 A resolution from electron micrograph | | Descriptor: | Aquaporin-4 | | Authors: | Mitsuma, T, Tani, K, Hiroaki, Y, Kamegawa, A, Suzuki, H, Hibino, H, Kurachi, Y, Fujiyoshi, Y. | | Deposit date: | 2010-07-24 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Influence of the cytoplasmic domains of aquaporin-4 on water conduction and array formation.
J.Mol.Biol., 402, 2010
|
|
2ZZ9
 
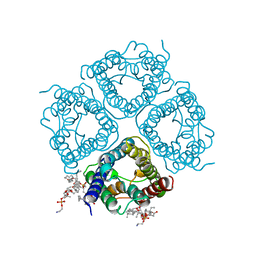 | | Structure of aquaporin-4 S180D mutant at 2.8 A resolution by electron crystallography | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Aquaporin-4 | | Authors: | Tani, K, Mitsuma, T, Hiroaki, Y, Kamegawa, A, Nishikawa, K, Tanimura, Y, Fujiyoshi, Y. | | Deposit date: | 2009-02-06 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Mechanism of Aquaporin-4's Fast and Highly Selective Water Conduction and Proton Exclusion.
J.Mol.Biol., 389, 2009
|
|
5NNW
 
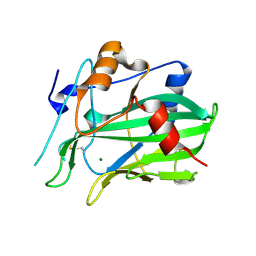 | | NLPPya in complex with glucosamine | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, 25 kDa protein elicitor, MAGNESIUM ION | | Authors: | Podobnik, M, Anderluh, G, Lenarcic, T. | | Deposit date: | 2017-04-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins.
Science, 358, 2017
|
|
5NO9
 
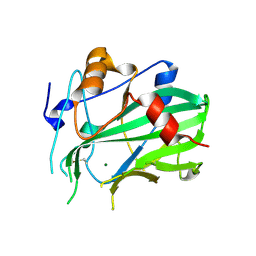 | | NLPPya in complex with mannosamine | | Descriptor: | 2-amino-2-deoxy-alpha-D-mannopyranose, 25 kDa protein elicitor, MAGNESIUM ION | | Authors: | Podobnik, M, Anderluh, G, Lenarcic, T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins.
Science, 358, 2017
|
|
8IF3
 
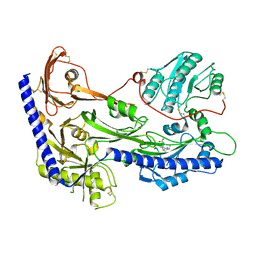 | | Structure of human alpha-2/delta-1 with mirogabalin | | Descriptor: | 2-[(1R,5S,6S)-6-(aminomethyl)-3-ethyl-6-bicyclo[3.2.0]hept-3-enyl]acetic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kozai, D, Numoto, N, Fujiyoshi, Y. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Recognition Mechanism of a Novel Gabapentinoid Drug, Mirogabalin, for Recombinant Human alpha 2 delta 1, a Voltage-Gated Calcium Channel Subunit.
J.Mol.Biol., 435, 2023
|
|
8IF4
 
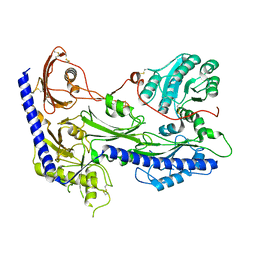 | | Structure of human alpha-2/delta-1 without mirogabalin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Voltage-dependent calcium channel subunit alpha-2/delta-1 | | Authors: | Kozai, D, Numoto, N, Fujiyoshi, Y. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Recognition Mechanism of a Novel Gabapentinoid Drug, Mirogabalin, for Recombinant Human alpha 2 delta 1, a Voltage-Gated Calcium Channel Subunit.
J.Mol.Biol., 435, 2023
|
|
7PUD
 
 | | Bryoporin - actinoporin from moss Physcomitrium patens | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bryoporin, SULFATE ION | | Authors: | Solinc, G, Anderluh, G, Podobnik, M. | | Deposit date: | 2021-09-29 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Pore-forming moss protein bryoporin is structurally and mechanistically related to actinoporins from evolutionarily distant cnidarians.
J.Biol.Chem., 298, 2022
|
|
4ITF
 
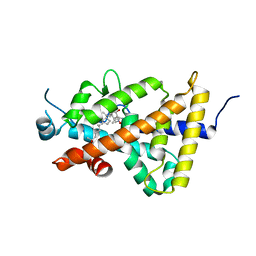 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 1alpha,25-Dihydroxy-2alpha-[2-(1H-tetrazole-1-yl)ethyl]vitamin D3 | | Descriptor: | (1R,2S,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-methyl-6-oxidanyl-heptan-2-yl]-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-2-[2-(1,2,3,4-tetrazol-1-yl)ethyl]cyclohexane-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2013-01-18 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Synthesis of 2 alpha-heteroarylalkyl active vitamin d3 with therapeutic effect on enhancing bone mineral density in vivo
ACS MED.CHEM.LETT., 4, 2013
|
|
4ITE
 
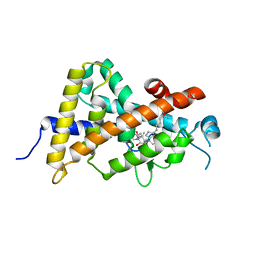 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 1alpha,25-Dihydroxy-2alpha-[2-(2H-tetrazol-2-yl)ethyl]vitamin D3 | | Descriptor: | (1R,2S,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-methyl-6-oxidanyl-heptan-2-yl]-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-2-[2-(1,2,3,4-tetrazol-2-yl)ethyl]cyclohexane-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2013-01-18 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Synthesis of 2 alpha-heteroarylalkyl active vitamin d3 with therapeutic effect on enhancing bone mineral density in vivo
ACS MED.CHEM.LETT., 4, 2013
|
|
8T8A
 
 | |
