1F8Y
 
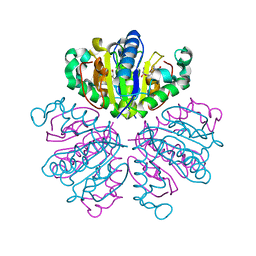 | | CRYSTAL STRUCTURE ANALYSIS OF NUCLEOSIDE 2-DEOXYRIBOSYLTRANSFERASE COMPLEXED WITH 5-METHYL-2'-DEOXYPSEUDOURIDINE | | Descriptor: | 2'-deoxy-1-methyl-pseudouridine, NUCLEOSIDE 2-DEOXYRIBOSYLTRANSFERASE | | Authors: | Armstrong, S.R, Cook, W.J, Short, S.A, Ealick, S.E. | | Deposit date: | 2000-07-05 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of nucleoside 2-deoxyribosyltransferase in native and ligand-bound forms reveal architecture of the active site.
Structure, 4, 1996
|
|
1FG9
 
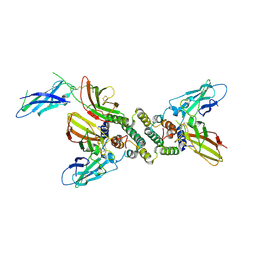 | | 3:1 COMPLEX OF INTERFERON-GAMMA RECEPTOR WITH INTERFERON-GAMMA DIMER | | Descriptor: | INTERFERON GAMMA, INTERFERON-GAMMA RECEPTOR ALPHA CHAIN | | Authors: | Thiel, D.J, le Du, M.-H, Walter, R.L, D'Arcy, A, Chene, C, Fountoulakis, M, Garotta, G, Winkler, F.K, Ealick, S.E. | | Deposit date: | 2000-07-28 | | Release date: | 2000-08-11 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Observation of an unexpected third receptor molecule in the crystal structure of human interferon-gamma receptor complex.
Structure Fold.Des., 8, 2000
|
|
1J58
 
 | | Crystal Structure of Oxalate Decarboxylase | | Descriptor: | FORMIC ACID, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Anand, R, Dorrestein, P.C, Kinsland, C, Begley, T.P, Ealick, S.E. | | Deposit date: | 2002-02-25 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 A resolution.
Biochemistry, 41, 2002
|
|
1JEN
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE | | Descriptor: | PROTEIN (S-ADENOSYLMETHIONINE DECARBOXYLASE (ALPHA CHAIN)), PROTEIN (S-ADENOSYLMETHIONINE DECARBOXYLASE (BETA CHAIN)) | | Authors: | Ekstrom, J.L, Mathews, I.I, Stanley, B.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 1999-02-23 | | Release date: | 1999-06-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of human S-adenosylmethionine decarboxylase at 2.25 A resolution reveals a novel fold.
Structure Fold.Des., 7, 1999
|
|
1JXH
 
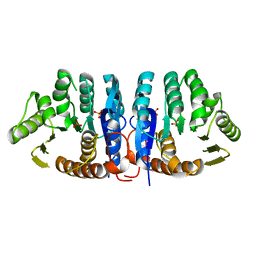 | | 4-Amino-5-hydroxymethyl-2-methylpyrimidine Phosphate Kinase from Salmonella typhimurium | | Descriptor: | PHOSPHOMETHYLPYRIMIDINE KINASE, SULFATE ION | | Authors: | Cheng, G, Bennett, E.M, Begley, T.P, Ealick, S.E. | | Deposit date: | 2001-09-07 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase from Salmonella typhimurium at 2.3 A resolution.
Structure, 10, 2002
|
|
1JXI
 
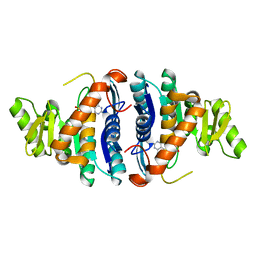 | | 4-Amino-5-hydroxymethyl-2-methylpyrimidine Phosphate Kinase from Salmonella typhimurium complexed with 4-Amino-5-hydroxymethyl-2-methylpyrimidine | | Descriptor: | 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, PHOSPHOMETHYLPYRIMIDINE KINASE, SULFATE ION | | Authors: | Cheng, G, Bennett, E.M, Begley, T.P, Ealick, S.E. | | Deposit date: | 2001-09-07 | | Release date: | 2002-02-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase from Salmonella typhimurium at 2.3 A resolution.
Structure, 10, 2002
|
|
1K3F
 
 | | Uridine Phosphorylase from E. coli, Refined in the Monoclinic Crystal Lattice | | Descriptor: | uridine phosphorylase | | Authors: | Morgunova, E.Yu, Mikhailov, A.M, Popov, A.N, Blagova, E.V, Smirnova, E.A, Vainshtein, B.K, Mao, C, Armstrong, S.R, Ealick, S.E, Komissarov, A.A, Linkova, E.V, Burlakova, A.A, Mironov, A.S, Debabov, V.G. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structure at 2.5 A resolution of uridine phosphorylase from E. coli as refined in the monoclinic crystal lattice.
FEBS Lett., 367, 1995
|
|
2OP2
 
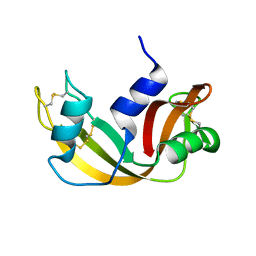 | |
1SJ9
 
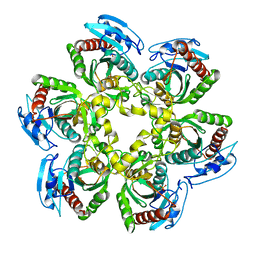 | | Crystal structure of the uridine phosphorylase from Salmonella typhimurium at 2.5A resolution | | Descriptor: | PHOSPHATE ION, Uridine phosphorylase | | Authors: | Dontsova, M, Gabdoulkhakov, A, Morgunova, E, Garber, M, Nikonov, S, Betzel, C, Ealick, S, Mikhailov, A. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preliminary investigation of the three-dimensional structure of Salmonella typhimurium uridine phosphorylase in the crystalline state.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
5CLG
 
 | |
5CLJ
 
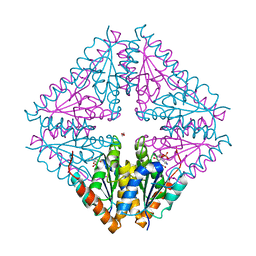 | |
1UW8
 
 | | CRYSTAL STRUCTURE OF OXALATE DECARBOXYLASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Stevenson, C.E.M, Bowater, L, Tanner, A, Lawson, D.M, Bornemann, S. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Closed Conformation of Bacillus Subtilis Oxalate Decarboxylase Oxdc Provides Evidence for the True Identity of the Active Site
J.Biol.Chem., 279, 2004
|
|
2RD3
 
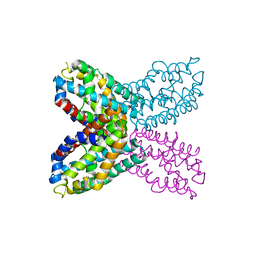 | |
3SXP
 
 | |
1A3Z
 
 | | REDUCED RUSTICYANIN AT 1.9 ANGSTROMS | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Zhao, D, Shoham, M. | | Deposit date: | 1998-01-27 | | Release date: | 1998-07-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rusticyanin: Extremes in acid stability and redox potential explained by the crystal structure.
Biophys.J., 74, 1998
|
|
1A8Z
 
 | | STRUCTURE DETERMINATION OF A 16.8KDA COPPER PROTEIN RUSTICYANIN AT 2.1A RESOLUTION USING ANOMALOUS SCATTERING DATA WITH DIRECT METHODS | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Harvey, I, Hao, Q, Duke, E.M.H, Ingledew, W.J, Hasnain, S.S. | | Deposit date: | 1998-03-30 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of a 16.8 kDa copper protein at 2.1 A resolution using anomalous scattering data with direct methods.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1B8N
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-1-(S)-(9-DEAZAGUANIN-9-YL)-D-RIBITOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fedorov, A.A, Kicska, G.A, Fedorov, E.V, Strokopytov, B.V, Tyler, P.C, Furneaux, R.H, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-02-02 | | Release date: | 1999-02-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic dissection of the hydrogen bond network for transition-state analogue binding to purine nucleoside phosphorylase
Biochemistry, 41, 2002
|
|
1B8O
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fedorov, A.A, Kicska, G.A, Fedorov, E.V, Shi, W, Tyler, P.C, Furneaux, R.H, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-02-02 | | Release date: | 1999-02-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis.
Biochemistry, 40, 2001
|
|
3FE5
 
 | | Crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from bovine kidney | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (III) ION | | Authors: | Dilovic, I, Gliubich, F, Malpeli, G, Zanotti, G, Matkovic-Calogovic, D. | | Deposit date: | 2008-11-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of bovine 3-hydroxyanthranilate 3,4-dioxygenase.
Biopolymers, 2009
|
|
3HPE
 
 | |
3IBX
 
 | | Crystal structure of F47Y variant of TenA (HP1287) from Helicobacter pylori | | Descriptor: | Putative thiaminase II | | Authors: | Barison, N, Cendron, L, Trento, A, Angelini, A, Zanotti, G. | | Deposit date: | 2009-07-17 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mutational analysis of TenA protein (HP1287) from the Helicobacter pylori thiamin salvage pathway - evidence of a different substrate specificity.
Febs J., 276, 2009
|
|
1RCT
 
 | | Crystal structure of Human purine nucleoside phosphorylase complexed with INOSINE | | Descriptor: | INOSINE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Canduri, F, dos Santos, D.M, Silva, R.G, Mendes, M.A, Palma, M.S, de Azevedo Jr, W.F, Basso, L.A, Santos, D.S. | | Deposit date: | 2003-11-04 | | Release date: | 2004-01-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of human purine nucleoside phosphorylase complexed with inosine and ddI
Biochem.Biophys.Res.Commun., 313, 2004
|
|
1M73
 
 | | CRYSTAL STRUCTURE OF HUMAN PNP AT 2.3A RESOLUTION | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | De Azevedo Jr, W.F, Marangoni Dos Santos, D, Canduri, F, Santos, G.C, Olivieri, J.R, Silva, R.G, Basso, L.A, Palma, M.S, Santos, D.S. | | Deposit date: | 2002-07-18 | | Release date: | 2003-09-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human purine nucleoside phosphorylase at 2.3A resolution.
Biochem.Biophys.Res.Commun., 308, 2003
|
|
1SNB
 
 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M8 | | Descriptor: | NEUROTOXIN BMK M8 | | Authors: | Wang, D.C, Zeng, Z.H, Li, H.M. | | Deposit date: | 1997-03-12 | | Release date: | 1997-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an acidic neurotoxin from scorpion Buthus martensii Karsch at 1.85 A resolution.
J.Mol.Biol., 261, 1996
|
|
1SN4
 
 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M4 | | Descriptor: | ACETATE ION, PROTEIN (NEUROTOXIN BMK M4) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
